| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,069,619 – 17,069,715 |
| Length | 96 |
| Max. P | 0.797485 |

| Location | 17,069,619 – 17,069,715 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -25.48 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17069619 96 + 22407834 UGGAGCUAAAGCUGAAGCUGGAACCUGGCGGUGUAUCUGCUGGAUAUCCCACCUUGGCAUUAGCAUAACCAUUGCCAUUUCGGGUGAUGGGAAAUC ....(((((.((..(.(..(((..(..((((.....))))..)...)))..).)..)).)))))....((((..((......))..))))...... ( -34.30) >DroSec_CAF1 33646 90 + 1 UUGUC------CUGAAGCUGGAGCCUGGCGGUGUUCCUGCUGGAUAUCCCACCUUGGCAUUAGCAUAACCAUUGCCGUUUCGGGUGAUGGGAAAUC .....------.....(((((.(((.((.((..((((....))).)..)).))..))).)))))....((((..((......))..))))...... ( -31.70) >DroSim_CAF1 28941 90 + 1 UGGUC------CUGAAGCUGGAGCCUGGCGGUGUUCCUGCUGGAUAUCCCACCUUGGCAUUAGCAUAACCAUUGCCGUUUCGGGUGAUGGGAAAUC .....------.....(((((.(((.((.((..((((....))).)..)).))..))).)))))....((((..((......))..))))...... ( -31.70) >DroYak_CAF1 32562 89 + 1 UGGUC------CUGGACCUGGA-CCUGGCGGUGGUCCUGCUGGAUAUCCCACUUUGGCAUUAGCAUAACCAUUGCCAUUUCGGGUGAUGGGAAAUC ...((------(((.(((((((-..((((((((((..((((((.....((.....))..))))))..)))))))))).)))))))..))))).... ( -35.70) >consensus UGGUC______CUGAAGCUGGAGCCUGGCGGUGUUCCUGCUGGAUAUCCCACCUUGGCAUUAGCAUAACCAUUGCCAUUUCGGGUGAUGGGAAAUC ................(((((.(((.((.((.(.(((....)))...))).))..))).)))))....((((..((......))..))))...... (-25.48 = -26.10 + 0.62)

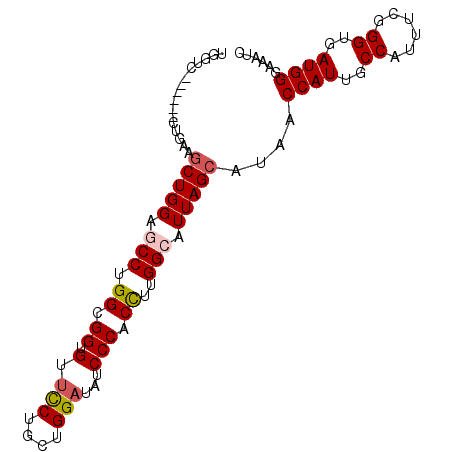

| Location | 17,069,619 – 17,069,715 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -20.50 |
| Energy contribution | -21.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17069619 96 - 22407834 GAUUUCCCAUCACCCGAAAUGGCAAUGGUUAUGCUAAUGCCAAGGUGGGAUAUCCAGCAGAUACACCGCCAGGUUCCAGCUUCAGCUUUAGCUCCA ...........(((.....(((((.(((.....))).))))).(((((..((((.....))))..))))).)))...((((........))))... ( -26.70) >DroSec_CAF1 33646 90 - 1 GAUUUCCCAUCACCCGAAACGGCAAUGGUUAUGCUAAUGCCAAGGUGGGAUAUCCAGCAGGAACACCGCCAGGCUCCAGCUUCAG------GACAA (((.(((((((....(...)((((.(((.....))).))))..))))))).))).(((.(((...((....)).))).)))....------..... ( -25.70) >DroSim_CAF1 28941 90 - 1 GAUUUCCCAUCACCCGAAACGGCAAUGGUUAUGCUAAUGCCAAGGUGGGAUAUCCAGCAGGAACACCGCCAGGCUCCAGCUUCAG------GACCA (((.(((((((....(...)((((.(((.....))).))))..))))))).)))..((.((....))))..((.(((.......)------)))). ( -25.90) >DroYak_CAF1 32562 89 - 1 GAUUUCCCAUCACCCGAAAUGGCAAUGGUUAUGCUAAUGCCAAAGUGGGAUAUCCAGCAGGACCACCGCCAGG-UCCAGGUCCAG------GACCA (((.(((((((....))..(((((.(((.....))).)))))...))))).))).....(((((.......))-))).((((...------)))). ( -30.80) >consensus GAUUUCCCAUCACCCGAAACGGCAAUGGUUAUGCUAAUGCCAAGGUGGGAUAUCCAGCAGGAACACCGCCAGGCUCCAGCUUCAG______GACCA (((.(((((((.........((((.(((.....))).))))..))))))).))).(((.(((...((....)).))).)))............... (-20.50 = -21.50 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:40 2006