
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,068,951 – 17,069,052 |
| Length | 101 |
| Max. P | 0.691879 |

| Location | 17,068,951 – 17,069,052 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
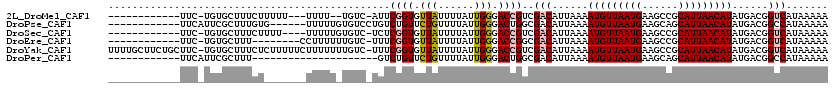
>2L_DroMel_CAF1 17068951 101 + 22407834 ------------UUC-UGUGCUUUCUUUUU---UUUU--UGUC-AUUCGGUGUUAUUUUAUUGGGACCGUCGACAUUAAAAUGUUAAUGAAGCCGCAUUAACAUAUGACGGUCAUAAAAA ------------...-((((((.((....(---((..--((((-...((((.(((......))).))))..))))..)))(((((((((......)))))))))..)).)).)))).... ( -24.10) >DroPse_CAF1 20305 102 + 1 ------------UUCAUUCGCUUUGUG------UUUUUGUGUCCUGUCUGUUCUGUUUUAUUGGGACUGGCGACAUUAAAAUGUUAAUGAAGCAGCAUUAACAUAUGACGGCCAUAAAAA ------------...............------((((((((.((((((.((((((......)))))).))))........(((((((((......))))))))).....)).)))))))) ( -23.60) >DroSec_CAF1 32956 102 + 1 ------------UUC-UGUGCUUUCUUUU----UUUUUGUGUC-UCUCGGUGUUAUUUUAUUGGGACCGUCGACAUUAAAAUGUUAAUGAAGCCGCAUUAACAUAUGACGGUCAUAAAAA ------------...-((((((.((....----.(((.(((((-...((((.(((......))).))))..))))).)))(((((((((......)))))))))..)).)).)))).... ( -24.60) >DroEre_CAF1 32505 98 + 1 ------------UUC-UGUGCUUU--------CCUUUUUUGUC-UUUCGGUGUUAUUUUAUUGGGACCGGCGACAUUAAAAUGUUAAUGAAGCCGCAUUAACAUAUGACGGUCAUAAAAA ------------...-((((((.(--------(.(((..((((-..(((((.(((......))).))))).))))..)))(((((((((......)))))))))..)).)).)))).... ( -23.70) >DroYak_CAF1 31764 118 + 1 UUUUGCUUCUGCUUC-UGUGCUUUCUCUUUUUCUUUUUUUGUC-UUUCGGUGUUAUUUUAUUGGGACCGUCGACAUUAAAAUGUUAAUGAAGCCGCAUUAACAUAUGACGGUCAUAAAAA ....((....))...-((((((.((.........(((..((((-...((((.(((......))).))))..))))..)))(((((((((......)))))))))..)).)).)))).... ( -25.10) >DroPer_CAF1 18354 87 + 1 ------------UUCAUUCGCUUU---------------------GUCUGUUCUGUUUUAUUGGGACUGGCGACAUUAAAAUGUUAAUGAAGCAGCAUUAACAUAUGACGGCCAUAAAAA ------------.......(((((---------------------(((.((((((......)))))).))))).......(((((((((......))))))))).....)))........ ( -19.00) >consensus ____________UUC_UGUGCUUUCU_______UUUUU_UGUC_UUUCGGUGUUAUUUUAUUGGGACCGGCGACAUUAAAAUGUUAAUGAAGCCGCAUUAACAUAUGACGGUCAUAAAAA ...............................................((((.(((......))).))))..(((......(((((((((......)))))))))......)))....... (-14.39 = -14.62 + 0.22)

| Location | 17,068,951 – 17,069,052 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -17.53 |
| Consensus MFE | -12.56 |
| Energy contribution | -12.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
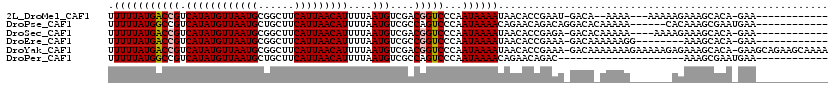
>2L_DroMel_CAF1 17068951 101 - 22407834 UUUUUAUGACCGUCAUAUGUUAAUGCGGCUUCAUUAACAUUUUAAUGUCGACGGUCCCAAUAAAAUAACACCGAAU-GACA--AAAA---AAAAAGAAAGCACA-GAA------------ .(((((((((((((..(((((((((......))))))))).........)))))))...))))))...........-....--....---..............-...------------ ( -17.80) >DroPse_CAF1 20305 102 - 1 UUUUUAUGGCCGUCAUAUGUUAAUGCUGCUUCAUUAACAUUUUAAUGUCGCCAGUCCCAAUAAAACAGAACAGACAGGACACAAAAA------CACAAAGCGAAUGAA------------ ......((((.(.((((((((((((......)))))))))....))).)))))((((...................)))).......------...............------------ ( -17.71) >DroSec_CAF1 32956 102 - 1 UUUUUAUGACCGUCAUAUGUUAAUGCGGCUUCAUUAACAUUUUAAUGUCGACGGUCCCAAUAAAAUAACACCGAGA-GACACAAAAA----AAAAGAAAGCACA-GAA------------ .(((((((((((((..(((((((((......))))))))).........)))))))...))))))...........-..........----.............-...------------ ( -17.80) >DroEre_CAF1 32505 98 - 1 UUUUUAUGACCGUCAUAUGUUAAUGCGGCUUCAUUAACAUUUUAAUGUCGCCGGUCCCAAUAAAAUAACACCGAAA-GACAAAAAAGG--------AAAGCACA-GAA------------ .(((((((((((.(((......))))(((..((((((....))))))..)))))))...))))))......(....-)..........--------........-...------------ ( -17.10) >DroYak_CAF1 31764 118 - 1 UUUUUAUGACCGUCAUAUGUUAAUGCGGCUUCAUUAACAUUUUAAUGUCGACGGUCCCAAUAAAAUAACACCGAAA-GACAAAAAAAGAAAAAGAGAAAGCACA-GAAGCAGAAGCAAAA .(((((((((((((..(((((((((......))))))))).........)))))))...))))))......(....-).....................((...-...)).......... ( -20.20) >DroPer_CAF1 18354 87 - 1 UUUUUAUGGCCGUCAUAUGUUAAUGCUGCUUCAUUAACAUUUUAAUGUCGCCAGUCCCAAUAAAACAGAACAGAC---------------------AAAGCGAAUGAA------------ ......((((.(.((((((((((((......)))))))))....))).)))))(((................)))---------------------............------------ ( -14.59) >consensus UUUUUAUGACCGUCAUAUGUUAAUGCGGCUUCAUUAACAUUUUAAUGUCGACGGUCCCAAUAAAAUAACACCGAAA_GACA_AAAAA_______AGAAAGCACA_GAA____________ .(((((((((((.((((((((((((......)))))))))....)))....)))))...))))))....................................................... (-12.56 = -12.67 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:38 2006