| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,817,552 – 1,817,663 |
| Length | 111 |
| Max. P | 0.750279 |

| Location | 1,817,552 – 1,817,663 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.45 |
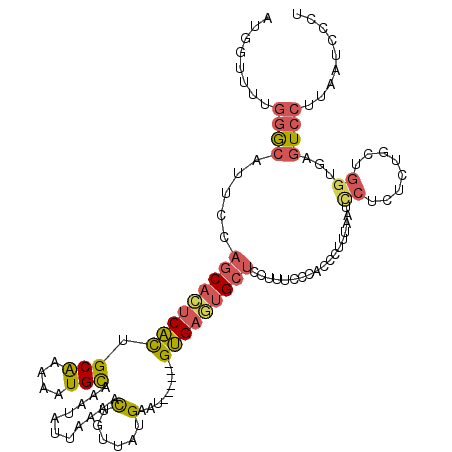
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
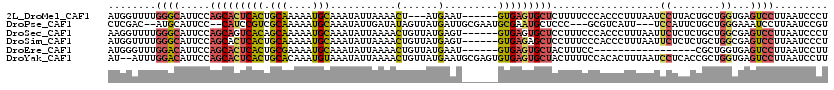
>2L_DroMel_CAF1 1817552 111 - 22407834 AUGGUUUUGGGCAUUCCAGCACUCACUGCAAAAAUGCAAAUAUUAAAACU---AUGAAU------GUGAGUGCUCUUUUCCCACCCUUUAAUCCUUACUGCUGGUGAGUCCUUAAUCCCU ..((((..((((.....(((((((((((((....))))....(((.....---.)))..------))))))))).......((((.................)))).))))..))))... ( -27.23) >DroPse_CAF1 3905 110 - 1 CUCGAC--AUGCAUUCC--CAUCCGUCGCAAAAAUGCAAAUAUUGAUAUAGUUAUGAUUGCGAAUGCGAAUGCUCCC---GCGUCAUU---UCCAUUCUGCUGGGAAAUCCUUAAUCCGU ......--..((((((.--(((...((((((..(((....(((....)))..)))..))))))))).))))))((((---(((.....---.......))).)))).............. ( -21.90) >DroSec_CAF1 37502 114 - 1 AAGGUUUUGGGCAUUCCAGCAGUCACAGCAAAAAUGCAAAUAUUAAAACUGUUAUGAGU------GUGAGUGCUCCUUUCCCACCCUUUAAUUCUCUCUGCUGGCGAGUCCUUAAUCCCU ..((((..((((...(((((((((((.(((....)))..........(((......)))------))))(((.........))).............)))))))...))))..))))... ( -27.10) >DroSim_CAF1 15154 114 - 1 AUGGUUUUGGGCAUUCCAGCACUCACUGCAAAAAUGCAAAUAUUAAAACUGUUAUGAGU------GUGAGAGCUCCUUUCCCACCCUUUAAUUCUCUCUGCUGGCGAGUCCUUAAUCCCU ..((((..((((...((((((((((.((((....))))((((.......)))).)))))------..(((((.....................))))).)))))...))))..))))... ( -26.90) >DroEre_CAF1 26018 97 - 1 AUGGGUUUGGACAUUCCAGCACUCACUGCGAAAAUGCAAAUAUUAAAACUGUUAUGAAU------GUGAGUGCUACUUUCC-----------------CGCUGGUGAGUCCUUAAUCCUU ..(((((.((((.....((((((((((((......)))((((.......))))......------)))))))))....(((-----------------(...)).))))))..))))).. ( -27.00) >DroYak_CAF1 2306 118 - 1 AU--AUUUGGACAUUCCAGCACUCACUGCACAAAUGUAAAUAUUAAAACUGUUAUGAAUGCGAGUGUGAGUGCUACUUUUCCACACUUUAAUCCUCACCGCUGGUGAGUCCUUAAUCCUU ..--....((((.....(((((((((((((....)))).........(((((.......)).))))))))))))....................(((((...)))))))))......... ( -27.80) >consensus AUGGUUUUGGGCAUUCCAGCACUCACUGCAAAAAUGCAAAUAUUAAAACUGUUAUGAAU______GUGAGUGCUCCUUUCCCACCCUUUAAUCCUCUCUGCUGGUGAGUCCUUAAUCCCU ........((((.....(((((((((.(((....)))...........(......).........)))))))))..................((........))...))))......... (-13.57 = -13.80 + 0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:49 2006