
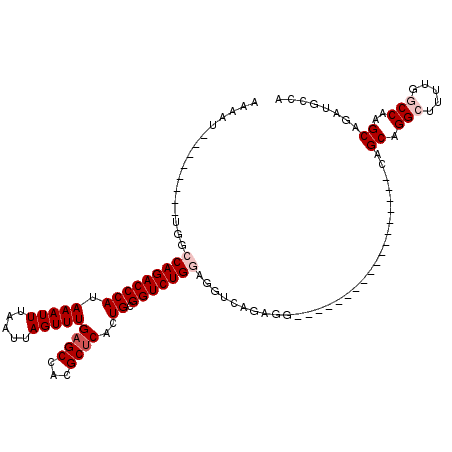
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,055,732 – 17,055,826 |
| Length | 94 |
| Max. P | 0.702381 |

| Location | 17,055,732 – 17,055,826 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.85 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17055732 94 + 22407834 AAAAU--------UGGCCAGACCCAUAAAUUUAAUUAGUUUGAGCCACGCUCACUGCGGUCUGGAGGUCAGAGG------------------CAGCAGGCUUUUGGCCAAGCAGAUGGCA .....--------(((((((((...............))))).))))........((.(((((..(((((((((------------------(.....))))))))))...))))).)). ( -33.76) >DroPse_CAF1 7963 117 + 1 AAAUCCAGAGACCCAGCCAGACCCAUAAAUUUAAUUAGUUUGAGCCACGCUCAGUGCGGUCUGAAGGUCAGUGGGAACACCAGCAGGAGCAGAAGCAGGCC---GUCCAAGCAGCUGCCA ...(((...((((....((((((((((((((.....)))))((((...)))).))).))))))..)))).(((....))).....)))((((..((.((..---..))..))..)))).. ( -35.70) >DroSec_CAF1 19795 94 + 1 AAAAU--------UGGCCAGACCCAUAAAUUUAAUUAGUUUGAGCCACGCUCACUGCGGUCUGGAGGUCAGAGG------------------CAGCAGGCUUUUGGCCAAGCAGAUGGCA .....--------(((((((((...............))))).))))........((.(((((..(((((((((------------------(.....))))))))))...))))).)). ( -33.76) >DroEre_CAF1 19588 94 + 1 AAAAU--------UGGCCAGACCCAUAAAUUUAAUUAGUUUGAGCCACGCCCACUGCGGUCUGGAGGUCAGAGG------------------CAGCAGGCUUUUGGCCAAGCAGAUGCCA .....--------((((((((((..((((((.....))))))......((.....))))))))..(((((((((------------------(.....))))))))))........)))) ( -31.20) >DroYak_CAF1 17560 94 + 1 AAAAU--------UGGCCAGACCCAUAAAUUUAAUUAGUUUGAGCCACGCUCACUGCGGUCUGGAGGUCAGAGG------------------CAGCAGGCUUUAGGCCAAGCAGAUGGCA .....--------(((((((((...............))))).))))........((.(((((..((((.((((------------------(.....))))).))))...))))).)). ( -29.96) >DroPer_CAF1 4101 117 + 1 AAAUCCAGAGACCCAGCCAGACCCAUAAAUUUAAUUAGUUUGAGCCACGCUCAGUGCGGUCUGAAGGUCAGUGGGAACACCAGCAGGAGCAGAAGCAGGCC---GUCCAAGCAGCUGCCA ...(((...((((....((((((((((((((.....)))))((((...)))).))).))))))..)))).(((....))).....)))((((..((.((..---..))..))..)))).. ( -35.70) >consensus AAAAU________UGGCCAGACCCAUAAAUUUAAUUAGUUUGAGCCACGCUCACUGCGGUCUGGAGGUCAGAGG__________________CAGCAGGCUUUUGGCCAAGCAGAUGCCA ................(((((((((.(((((.....)))))((((...))))..)).)))))))..............................((.(((.....)))..))........ (-18.02 = -18.85 + 0.83)

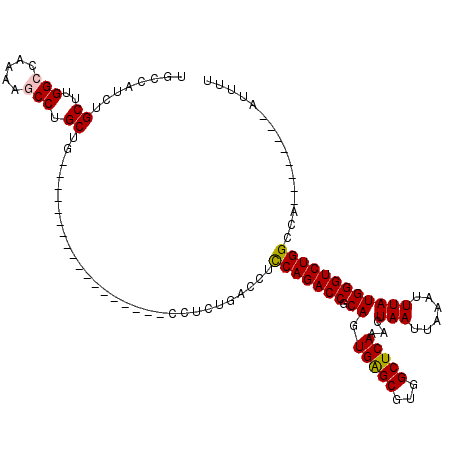
| Location | 17,055,732 – 17,055,826 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17055732 94 - 22407834 UGCCAUCUGCUUGGCCAAAAGCCUGCUG------------------CCUCUGACCUCCAGACCGCAGUGAGCGUGGCUCAAACUAAUUAAAUUUAUGGGUCUGGCCA--------AUUUU ..........(((((((...((..((((------------------(.((((.....))))..)))))..))..((((((...(((......)))))))))))))))--------).... ( -29.90) >DroPse_CAF1 7963 117 - 1 UGGCAGCUGCUUGGAC---GGCCUGCUUCUGCUCCUGCUGGUGUUCCCACUGACCUUCAGACCGCACUGAGCGUGGCUCAAACUAAUUAAAUUUAUGGGUCUGGCUGGGUCUCUGGAUUU .(((((..((..((..---..)).))..)))))((....)).....(((..((((((((((((.((.(((((...)))))...(((......))))))))))))..)))))..))).... ( -36.60) >DroSec_CAF1 19795 94 - 1 UGCCAUCUGCUUGGCCAAAAGCCUGCUG------------------CCUCUGACCUCCAGACCGCAGUGAGCGUGGCUCAAACUAAUUAAAUUUAUGGGUCUGGCCA--------AUUUU ..........(((((((...((..((((------------------(.((((.....))))..)))))..))..((((((...(((......)))))))))))))))--------).... ( -29.90) >DroEre_CAF1 19588 94 - 1 UGGCAUCUGCUUGGCCAAAAGCCUGCUG------------------CCUCUGACCUCCAGACCGCAGUGGGCGUGGCUCAAACUAAUUAAAUUUAUGGGUCUGGCCA--------AUUUU .(((....)))((((((...(((..(((------------------(.((((.....))))..))))..)))..((((((...(((......)))))))))))))))--------..... ( -36.40) >DroYak_CAF1 17560 94 - 1 UGCCAUCUGCUUGGCCUAAAGCCUGCUG------------------CCUCUGACCUCCAGACCGCAGUGAGCGUGGCUCAAACUAAUUAAAUUUAUGGGUCUGGCCA--------AUUUU ..........((((((....((..((((------------------(.((((.....))))..)))))..))..((((((...(((......))))))))).)))))--------).... ( -28.60) >DroPer_CAF1 4101 117 - 1 UGGCAGCUGCUUGGAC---GGCCUGCUUCUGCUCCUGCUGGUGUUCCCACUGACCUUCAGACCGCACUGAGCGUGGCUCAAACUAAUUAAAUUUAUGGGUCUGGCUGGGUCUCUGGAUUU .(((((..((..((..---..)).))..)))))((....)).....(((..((((((((((((.((.(((((...)))))...(((......))))))))))))..)))))..))).... ( -36.60) >consensus UGCCAUCUGCUUGGCCAAAAGCCUGCUG__________________CCUCUGACCUCCAGACCGCAGUGAGCGUGGCUCAAACUAAUUAAAUUUAUGGGUCUGGCCA________AUUUU ........((..(((.....))).))..............................(((((((.((.(((((...)))))...(((......))))))))))))................ (-19.48 = -19.45 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:28 2006