| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,051,299 – 17,051,742 |
| Length | 443 |
| Max. P | 0.999940 |

| Location | 17,051,299 – 17,051,402 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -22.09 |
| Energy contribution | -22.45 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051299 103 - 22407834 AAGCAGCGAUCCGACGCCAUAAAUGUUGCUCCUCCAACUCCGGCUCCAUAUCCUGC---UGAUCCUUUGGAGUCGGGCCAC--------------CAACUCGAGUGGCAGCCAAGAACUA ..((.(((......))).......((((......)))).(((((((((.(((....---.)))....)))))))))(((((--------------........))))).))......... ( -28.90) >DroSec_CAF1 15213 103 - 1 AAGCAGCGAUCUGACGCCAUAAAUGUUGCUCCUCCAACUCCGUCUCCAUAUCCUGC---UGCUCCUUUGGAGCUAGGCCAC--------------CAACUCGAGUGGCAACCAACAACUA .(((((.(((..((((........((((......))))..)))).....)))))))---)(((((...)))))...(((((--------------........)))))............ ( -27.00) >DroSim_CAF1 9496 103 - 1 AAGCAGCGAUCCGACGCCAUAAAUGUUUCUCCUCCAACUCCGGCUCCAUAUCCUGC---UGCUCCUUUGGAGCUGGGCCAC--------------CAACUCGAGUGGCAACCAACAACUA .(((((.(((..((.(((......(((........)))...)))))...)))))))---)(((((...)))))((((((((--------------........)))))..)))....... ( -28.10) >DroEre_CAF1 15084 117 - 1 AAGCAGCGAUCCGACGCCAUAAAUGUUGCUCCUCCAACUCGGGCUCCAUGCCCUGC---UGCUCCUUUGGAGUUGGGCCACCAGCCACCAGUCACCAGCUCGAGUGGCGACCGACAACUA .....(((......)))......(((((.((..(((.(((((((.....)))).((---((.....((((.(((((....)))))..))))....))))..)))))).)).))))).... ( -37.90) >DroYak_CAF1 13136 106 - 1 AAGCAGCGAUCCGACGCCAUAAAUGUUGCUCCUCCAACUCCGGCUCCAUGUCCUGCACUUGCUCCUUUGGAGCUGGGCCUC--------------CAACUCGAGUGGCAACCCACAACUA .....(((......))).......(((((..(((....((((((((((......((....)).....))))))))))....--------------......)))..)))))......... ( -30.24) >consensus AAGCAGCGAUCCGACGCCAUAAAUGUUGCUCCUCCAACUCCGGCUCCAUAUCCUGC___UGCUCCUUUGGAGCUGGGCCAC______________CAACUCGAGUGGCAACCAACAACUA .....(((......))).......(((((..(((....((((((((((......((....)).....))))))))))........................)))..)))))......... (-22.09 = -22.45 + 0.36)

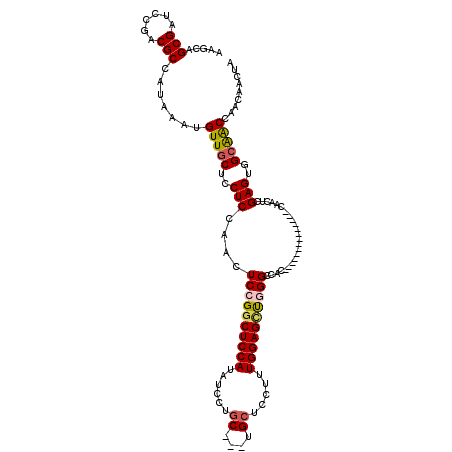
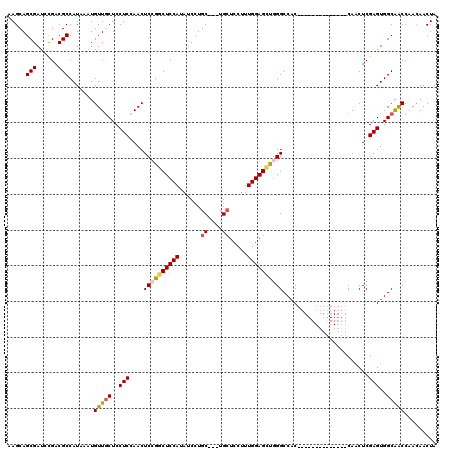
| Location | 17,051,402 – 17,051,514 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -35.84 |
| Energy contribution | -35.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051402 112 - 22407834 CAAAAAAAGGAGGUGGUGCACGGUGGAAG--------GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUGGUCAAAGCAUUGCGUCAGUCAAGUGGCACUCGGAACAGUU .........((((..(((((((((((...--------((((((((((((..........)))).)))))))).)))))..))....))))..)((((......)))).)))......... ( -37.80) >DroSec_CAF1 15316 112 - 1 CAAAAAAAGGGGGUGGUGCACGGUGGACG--------GGGUGACAGCCACAAAGACGCUUGGCUUGUCACCCGCUACUUGGUCAAAGCAUUGCGUCAGUCAAGUGGCACUCGGAACAGUU ..........(((((.(((..((..((((--------((((((((((((..........)))).))))))))(((..........)))....))))..))..))).)))))......... ( -37.80) >DroSim_CAF1 9599 112 - 1 CAAAAAAAGGGGGUGGUGCACGGUGGACG--------GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUGGUCAAAGCAUUGCGUCAGUCAAGUGGCACUCGGAACAGUU ..........(((((.(((..((..((((--------((((((((((((..........)))).))))))))(((..........)))....))))..))..))).)))))......... ( -37.80) >DroEre_CAF1 15201 119 - 1 CAAA-AAAGGGGGUGGUGCUCGGUGGACGGGUGGAGGGGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUGGUCAAAGCAUUGCGUCAGUCAAGUGGCACUCGGAACAGUU ....-...(((.(..(((((...((..(((((((...((((((((((((..........)))).)))))))).)))))))..)).)))))..)((((......)))).)))......... ( -46.90) >DroYak_CAF1 13242 112 - 1 CAAAAAAAGGGGGUGGUGCUCGGUAGACG--------GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUGGUCAAAGCAUUGCGUCAGUCAAGUGGCACUCGGAACAGUU ..........(((((.((((.((..((((--------((((((((((((..........)))).))))))))(((..........)))....))))..)).)))).)))))......... ( -40.20) >consensus CAAAAAAAGGGGGUGGUGCACGGUGGACG________GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUGGUCAAAGCAUUGCGUCAGUCAAGUGGCACUCGGAACAGUU .........(((.........................((((((((((((..........)))).))))))))(((((((((.....((...)).....))))))))).)))......... (-35.84 = -35.68 + -0.16)

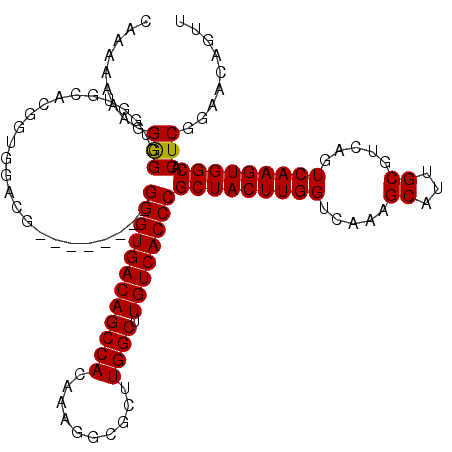
| Location | 17,051,442 – 17,051,544 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051442 102 + 22407834 CAAGUAGCGGGUGACAAGCCAAGCGCCUUUGUGGCUGUCACCC--------CUUCCACCGUGCACCACCUCCUUUUUUUGGCGGAGCUGC----------CUUCAACUCCACACAUAAGU ...(((((((((((((.((((..........))))))))))))--------..(((.(((..................))).))))))))----------.................... ( -32.17) >DroSec_CAF1 15356 102 + 1 CAAGUAGCGGGUGACAAGCCAAGCGUCUUUGUGGCUGUCACCC--------CGUCCACCGUGCACCACCCCCUUUUUUUGGCGGAGCUGC----------CUUCAACUCCACACAUAAGU ...(((((((((((((.(((..(((....))))))))))))))--------..(((.(((..................))).))))))))----------.................... ( -32.37) >DroSim_CAF1 9639 102 + 1 CAAGUAGCGGGUGACAAGCCAAGCGCCUUUGUGGCUGUCACCC--------CGUCCACCGUGCACCACCCCCUUUUUUUGGCGGAGCUGC----------CUUCAACUCCACACAUAAGU ...(((((((((((((.((((..........))))))))))))--------..(((.(((..................))).))))))))----------.................... ( -32.17) >DroEre_CAF1 15241 109 + 1 CAAGUAGCGGGUGACAAGCCAAGCGCCUUUGUGGCUGUCACCCCCUCCACCCGUCCACCGAGCACCACCCCCUUU-UUUGGCGAUGCUGC----------CUGCAACUCCACACAUAAGU ((.(((((((((((((.((((..........))))))))))))........((.(((..(((.........))).-..)))))..)))))----------.))................. ( -29.90) >DroYak_CAF1 13282 112 + 1 CAAGUAGCGGGUGACAAGCCAAGCGCCUUUGUGGCUGUCACCC--------CGUCUACCGAGCACCACCCCCUUUUUUUGGCGGUGCUGCCUUUCACUGCCUUCAACUCCACACAUAAGU ...(((((((((((((.((((..........))))))))).))--------)).))))...(((.((((.((.......)).)))).))).............................. ( -35.20) >consensus CAAGUAGCGGGUGACAAGCCAAGCGCCUUUGUGGCUGUCACCC________CGUCCACCGUGCACCACCCCCUUUUUUUGGCGGAGCUGC__________CUUCAACUCCACACAUAAGU ...(((((((((((((.((((..........))))))))))))........................((.((.......)).)).))))).............................. (-28.06 = -28.26 + 0.20)


| Location | 17,051,442 – 17,051,544 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -37.18 |
| Energy contribution | -37.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051442 102 - 22407834 ACUUAUGUGUGGAGUUGAAG----------GCAGCUCCGCCAAAAAAAGGAGGUGGUGCACGGUGGAAG--------GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUG ......(((((((((((...----------.)))))))((((...........))))))))(((((...--------((((((((((((..........)))).)))))))).))))).. ( -40.90) >DroSec_CAF1 15356 102 - 1 ACUUAUGUGUGGAGUUGAAG----------GCAGCUCCGCCAAAAAAAGGGGGUGGUGCACGGUGGACG--------GGGUGACAGCCACAAAGACGCUUGGCUUGUCACCCGCUACUUG .....((.(((((((((...----------.)))))))))))........(((((((...((.....))--------((((((((((((..........)))).))))))))))))))). ( -42.80) >DroSim_CAF1 9639 102 - 1 ACUUAUGUGUGGAGUUGAAG----------GCAGCUCCGCCAAAAAAAGGGGGUGGUGCACGGUGGACG--------GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUG .....((.(((((((((...----------.)))))))))))........(((((((...((.....))--------((((((((((((..........)))).))))))))))))))). ( -42.80) >DroEre_CAF1 15241 109 - 1 ACUUAUGUGUGGAGUUGCAG----------GCAGCAUCGCCAAA-AAAGGGGGUGGUGCUCGGUGGACGGGUGGAGGGGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUG ...........((((.((.(----------(((.((((.((...-...)).)))).)))).((..(((((((.(((..(....).(((.....))).))).)))))))..)))).)))). ( -43.40) >DroYak_CAF1 13282 112 - 1 ACUUAUGUGUGGAGUUGAAGGCAGUGAAAGGCAGCACCGCCAAAAAAAGGGGGUGGUGCUCGGUAGACG--------GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUG .....(((.(........).)))......((((.((((.((.......)).)))).)))).(((((...--------((((((((((((..........)))).)))))))).))))).. ( -42.50) >consensus ACUUAUGUGUGGAGUUGAAG__________GCAGCUCCGCCAAAAAAAGGGGGUGGUGCACGGUGGACG________GGGUGACAGCCACAAAGGCGCUUGGCUUGUCACCCGCUACUUG .....((.(((((((((..............)))))))))))........(((((((...((.....))........((((((((((((..........)))).))))))))))))))). (-37.18 = -37.46 + 0.28)

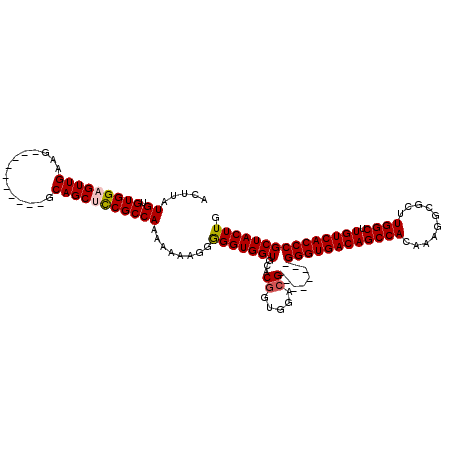
| Location | 17,051,482 – 17,051,584 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.01 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -30.50 |
| Energy contribution | -31.34 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051482 102 - 22407834 AAGCAACUUUGUUGCAGUCUCUGUGGCGCCAUGGAAGUGCACUUAUGUGUGGAGUUGAAG----------GCAGCUCCGCCAAAAAAAGGAGGUGGUGCACGGUGGAAG--------GGG ..(((((...)))))..((((((((.((((((..(((....))).((.(((((((((...----------.)))))))))))..........))))))))))).)))..--------... ( -36.30) >DroSec_CAF1 15396 102 - 1 AAGCAACUUUGUUGCAGUCUCUGUGGCCCCAUGGAAGUGCACUUAUGUGUGGAGUUGAAG----------GCAGCUCCGCCAAAAAAAGGGGGUGGUGCACGGUGGACG--------GGG ..(((((...))))).((((((((((((((....(((....))).((.(((((((((...----------.))))))))))).......)))))....))))).)))).--------... ( -39.00) >DroSim_CAF1 9679 102 - 1 AAGCAACUUUGUUGCAGUCUCUGUGGCCCCAUGGAAGUGCACUUAUGUGUGGAGUUGAAG----------GCAGCUCCGCCAAAAAAAGGGGGUGGUGCACGGUGGACG--------GGG ..(((((...))))).((((((((((((((....(((....))).((.(((((((((...----------.))))))))))).......)))))....))))).)))).--------... ( -39.00) >DroEre_CAF1 15281 109 - 1 AAGCAACUUUGUUGCACUCUCUGAGGCCCCAUGGAAGUGCACUUAUGUGUGGAGUUGCAG----------GCAGCAUCGCCAAA-AAAGGGGGUGGUGCUCGGUGGACGGGUGGAGGGGG ..(((((...))))).((((((...(((((.((.((.(.(((......))).).)).))(----------((......)))...-....)))))...(((((.....))))))))))).. ( -40.00) >DroYak_CAF1 13322 112 - 1 AAGCAACUUUGUUGCAGUCUCUGUGGCCCCAUGGAAGUGCACUUAUGUGUGGAGUUGAAGGCAGUGAAAGGCAGCACCGCCAAAAAAAGGGGGUGGUGCUCGGUAGACG--------GGG ..((...((..((((..((((((((....))))))...((((....))))......))..))))..))..))(((((((((..........)))))))))((.....))--------... ( -36.80) >consensus AAGCAACUUUGUUGCAGUCUCUGUGGCCCCAUGGAAGUGCACUUAUGUGUGGAGUUGAAG__________GCAGCUCCGCCAAAAAAAGGGGGUGGUGCACGGUGGACG________GGG ..(((((...))))).((((((((((((((....(((....))).((.(((((((((..............))))))))))).......)))))....))))).))))............ (-30.50 = -31.34 + 0.84)

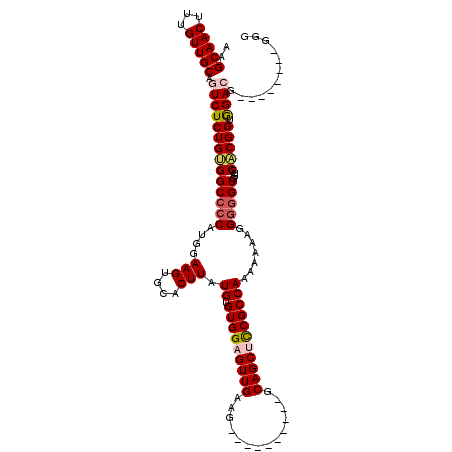
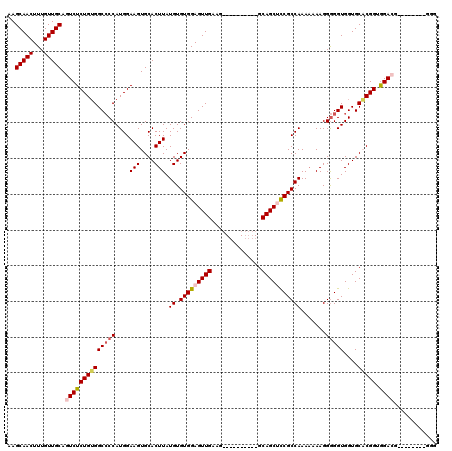
| Location | 17,051,584 – 17,051,704 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -47.14 |
| Consensus MFE | -42.38 |
| Energy contribution | -42.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.70 |
| SVM RNA-class probability | 0.999940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051584 120 + 22407834 UUGCCACGAAUGCGACAGUGGUUGCAAAUUCGAGCUCAGUUGCUCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGGUGCUUAAGGUGCCAGCUGAAGAGCUUAUUUCAUAAGGAGGUU ..(((.((((((((((....))))))..))))(((((.(((.(((....))))))((((((.(((.(((((((((....)))))))))))).))))))))))).............))). ( -48.50) >DroSec_CAF1 15498 120 + 1 UUGCCACGAAUGCGACAGUGGUUGCAAAUUCGAGCUCAGUUGCUUGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCAGCUGAAGAGCUUAUUCCAUAAGGAGGUU ..(((.((((((((((....))))))..))))(((((.(((.(((....))))))((((((.(((.(((((((((....)))))))))))).)))))))))))...(((....)))))). ( -47.70) >DroSim_CAF1 9781 120 + 1 UUGCCACGAAUGCGACAGUGGUUGCAAAUUCGAGCUCAGUUGCUCUCUGGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCAGCUGAAGAGCUUAUUUCAUAAGGAGGUU ..(((.((((((((((....))))))..))))(((((.(((.(((....))))))((((((.(((.(((((((((....)))))))))))).))))))))))).............))). ( -48.10) >DroEre_CAF1 15390 119 + 1 UUAUCACGAAUGCGACAGUGGUUGCAAAUUUGAGCUCAGUUGCGCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCGGCUGAAGAGCGUAUUUCAUAAGG-GGUU ......((((((((((....))))))..))))(((((...((.(((((......(((((((((((.(((((((((....)))))))))))))))))))))))))....))....)-)))) ( -48.40) >DroYak_CAF1 13434 120 + 1 UUACCACGAAUGCGACAGUGGUUGCAAAUUCGAGCUCAGUUGCGCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCGGCUUAAGAGGAUAUUUCAAAAGGAGGUU ..(((.((((((((((....))))))..))))(((.(......).))).((((.(((((((((((.(((((((((....))))))))))))))))...))))...)))).......))). ( -43.00) >consensus UUGCCACGAAUGCGACAGUGGUUGCAAAUUCGAGCUCAGUUGCUCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCAGCUGAAGAGCUUAUUUCAUAAGGAGGUU ..(((.((((((((((....))))))..)))).((((.(((.(((....))))))((((((.(((.(((((((((....)))))))))))).))))))))))..............))). (-42.38 = -42.46 + 0.08)


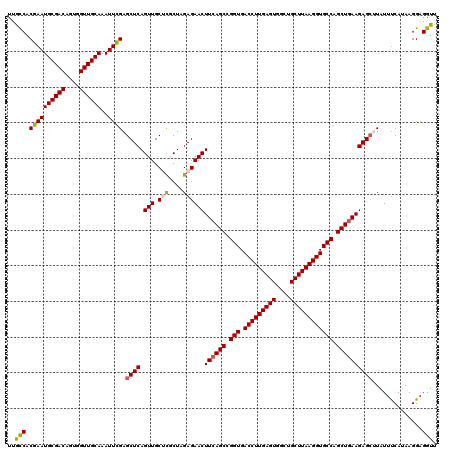
| Location | 17,051,584 – 17,051,704 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -30.82 |
| Energy contribution | -31.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051584 120 - 22407834 AACCUCCUUAUGAAAUAAGCUCUUCAGCUGGCACCUUAAGCACCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGAGCAACUGAGCUCGAAUUUGCAACCACUGUCGCAUUCGUGGCAA ..................((.((((((((((.(((((.((......)).)))))..))))))))))........(((((......))))).....))......((((((....)))))). ( -38.00) >DroSec_CAF1 15498 120 - 1 AACCUCCUUAUGGAAUAAGCUCUUCAGCUGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCAAGCAACUGAGCUCGAAUUUGCAACCACUGUCGCAUUCGUGGCAA ..........(((....((..((((((((((.(((((.((......)).)))))..))))))))))..))...((((((......)))......)))..))).((((((....)))))). ( -35.70) >DroSim_CAF1 9781 120 - 1 AACCUCCUUAUGAAAUAAGCUCUUCAGCUGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCCAGAGAGCAACUGAGCUCGAAUUUGCAACCACUGUCGCAUUCGUGGCAA ..........((((((.((((((((((((((.(((((.((......)).)))))..)))))))))(((((....)))))....)))))...)))).)).....((((((....)))))). ( -37.80) >DroEre_CAF1 15390 119 - 1 AACC-CCUUAUGAAAUACGCUCUUCAGCCGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGCGCAACUGAGCUCAAAUUUGCAACCACUGUCGCAUUCGUGAUAA ....-..(((((((....(((((((((((((.(((((.((......)).)))))..))))))))))......)))(((((.....((........))......)).))).)))))))... ( -36.30) >DroYak_CAF1 13434 120 - 1 AACCUCCUUUUGAAAUAUCCUCUUAAGCCGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGCGCAACUGAGCUCGAAUUUGCAACCACUGUCGCAUUCGUGGUAA ........(((((........(((.((((((.(((((.((......)).)))))..)))))).))).........((........)))))))......(((((..........))))).. ( -26.90) >consensus AACCUCCUUAUGAAAUAAGCUCUUCAGCUGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGAGCAACUGAGCUCGAAUUUGCAACCACUGUCGCAUUCGUGGCAA .......(((((((....(((((((((((((.(((((.((......)).)))))..)))))))))((((......))))....)))).......(((.((....)).))))))))))... (-30.82 = -31.46 + 0.64)

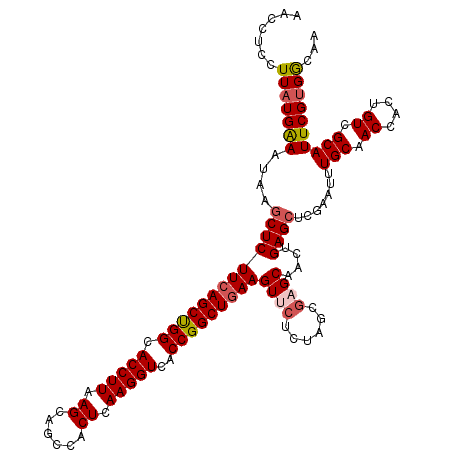
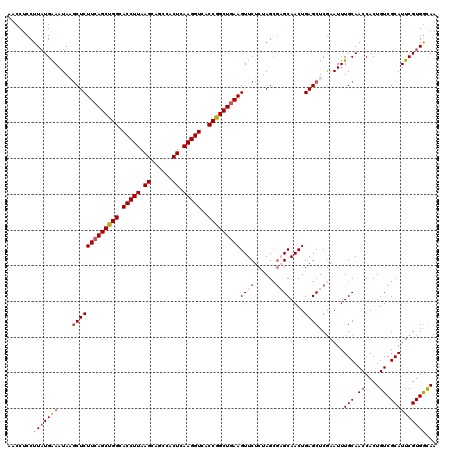
| Location | 17,051,624 – 17,051,742 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -33.06 |
| Energy contribution | -33.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051624 118 + 22407834 UGCUCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGGUGCUUAAGGUGCCAGCUGAAGAGCUUAUUUCAUAAGGAGGUUUUU--UUCUCGAGUGCCUUAUGUAUAAUAUAUAAAUAGUU .((((....(((..(((((((.(((.(((((((((....)))))))))))).)))))))..)))..........((((.....--.))))))))...(((((((...)))))))...... ( -39.30) >DroSec_CAF1 15538 120 + 1 UGCUUGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCAGCUGAAGAGCUUAUUCCAUAAGGAGGUUUUUUUUUCUCGAGUGCCCUAUGUAUAAUAUAUAAAUAGUU .((..(((.((((((((((((.(((.(((((((((....)))))))))))).)))))))......((((....))))........))))).))))).((((.((((...)))).)))).. ( -39.50) >DroSim_CAF1 9821 118 + 1 UGCUCUCUGGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCAGCUGAAGAGCUUAUUUCAUAAGGAGGUUUUUUUUUCUCGAGU--CCUAUGUAUAAUAUAUAAAUAGUU .((((....(((..(((((((.(((.(((((((((....)))))))))))).)))))))..)))..........((((........))))))))--.((((.((((...)))).)))).. ( -38.10) >DroEre_CAF1 15430 117 + 1 UGCGCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCGGCUGAAGAGCGUAUUUCAUAAGG-GGUUUUU--UUCUCGAGGGUCCUGUAUGUAAUAUAAAUAGGUUU ...(((((......(((((((((((.(((((((((....)))))))))))))))))))))))))(((.((((.((-((..(((--.....)))..)))).)))).)))............ ( -46.70) >DroYak_CAF1 13474 118 + 1 UGCGCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCGGCUUAAGAGGAUAUUUCAAAAGGAGGUUUUU--UUCUUGAGUGUCCUAUAUAAAAUUUAAAAAGGCUU .....(((......(((.(((((((.(((((((((....)))))))))))))))).)))(((((((((....((((((....)--))))))))))))))................))).. ( -41.70) >consensus UGCUCGCUAGAGAACUUCAGCCGGUGACCUUGAGUGGCUGCUUAAGGUGCCAGCUGAAGAGCUUAUUUCAUAAGGAGGUUUUU__UUCUCGAGUGCCCUAUGUAUAAUAUAUAAAUAGUU ....((((.((((((((((((.(((.(((((((((....)))))))))))).)))))))......((((....))))........))))).))))......................... (-33.06 = -33.62 + 0.56)

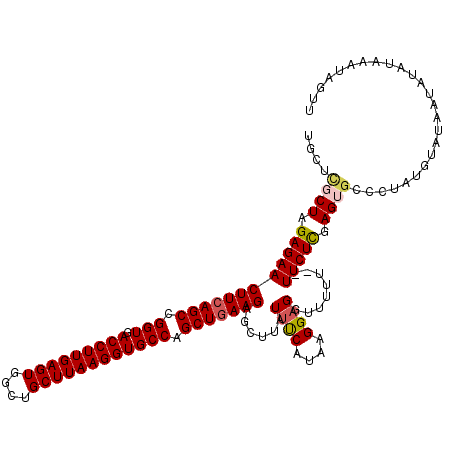
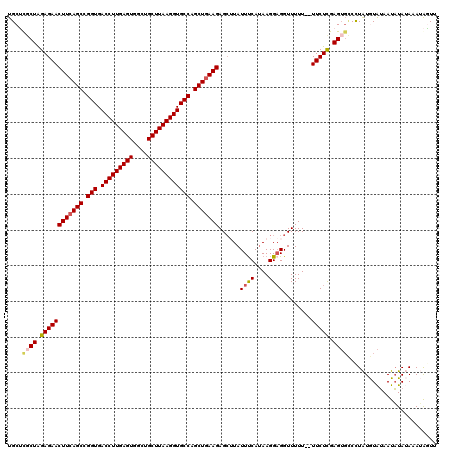
| Location | 17,051,624 – 17,051,742 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051624 118 - 22407834 AACUAUUUAUAUAUUAUACAUAAGGCACUCGAGAA--AAAAACCUCCUUAUGAAAUAAGCUCUUCAGCUGGCACCUUAAGCACCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGAGCA ..................(((((((..........--........)))))))......(((((((((((((.(((((.((......)).)))))..)))))))))((......)))))). ( -31.77) >DroSec_CAF1 15538 120 - 1 AACUAUUUAUAUAUUAUACAUAGGGCACUCGAGAAAAAAAAACCUCCUUAUGGAAUAAGCUCUUCAGCUGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCAAGCA ..((((.((((...)))).)))).((.((.(((((.........(((....))).......((((((((((.(((((.((......)).)))))..))))))))))))))).))...)). ( -33.80) >DroSim_CAF1 9821 118 - 1 AACUAUUUAUAUAUUAUACAUAGG--ACUCGAGAAAAAAAAACCUCCUUAUGAAAUAAGCUCUUCAGCUGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCCAGAGAGCA ..((((.((((...)))).)))).--.((((((((...........(((((...)))))..((((((((((.(((((.((......)).)))))..)))))))))))))))..))).... ( -31.10) >DroEre_CAF1 15430 117 - 1 AAACCUAUUUAUAUUACAUACAGGACCCUCGAGAA--AAAAACC-CCUUAUGAAAUACGCUCUUCAGCCGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGCGCA ...........((((.((((.(((...........--.......-))))))).))))((((((((((((((.(((((.((......)).)))))..))))))))))......)))).... ( -31.37) >DroYak_CAF1 13474 118 - 1 AAGCCUUUUUAAAUUUUAUAUAGGACACUCAAGAA--AAAAACCUCCUUUUGAAAUAUCCUCUUAAGCCGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGCGCA ..((((.....((((((....((((...((((((.--..........))))))....))))....((((((.(((((.((......)).)))))..))))))))))))....)).))... ( -25.50) >consensus AACUAUUUAUAUAUUAUACAUAGGACACUCGAGAA__AAAAACCUCCUUAUGAAAUAAGCUCUUCAGCUGGCACCUUAAGCAGCCACUCAAGGUCACCGGCUGAAGUUCUCUAGCGAGCA ...........................((.(((((...........(....).........((((((((((.(((((.((......)).)))))..))))))))))))))).))...... (-24.38 = -24.54 + 0.16)

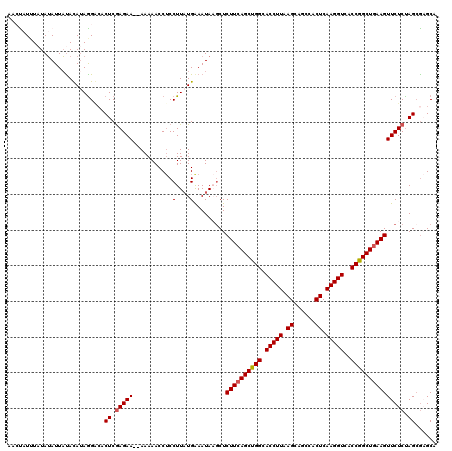
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:25 2006