
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,051,062 – 17,051,220 |
| Length | 158 |
| Max. P | 0.818121 |

| Location | 17,051,062 – 17,051,180 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -17.52 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051062 118 - 22407834 ACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAGGAUGCAAGUACAGUGAAAGCUCACAAAUAAAUUAAUGU-AUAUUUUUUUCAGAG-UU ..((((..(((....(((((((((..((.(....)))..)))))))))..((.(((....))).))....)))...))))(((((..(((.((((......-..)))).)))..)))-)) ( -23.50) >DroSec_CAF1 14976 117 - 1 ACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAAGAUGCAAGUACAGUGAAAACUCACAAAUAAAUACAUUU-UUAUUUGUUAGAAAA-A- ..((((..(((....(((((((((..((.(....)))..)))))))))...(((((.......)).))).)))...))))..((.(((((((((......)-)))))))).))....-.- ( -25.70) >DroSim_CAF1 9260 117 - 1 ACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAAGAUGCAACUACAGUGAAAACUCACAAAUAAAUGAAUGU-UUAUUUGUUAGAAAA-A- .......((((....(((((((((..((.(....)))..)))))))))...(((((.......)).)))......))))...((.((((((((((....))-)))))))).))....-.- ( -24.90) >DroEre_CAF1 14848 117 - 1 ACUUCAAUUACAACGCCUUCGGAGAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAACAGGAUGCCAGUGCAGUGAAAUCUCGCUAUAA-AGUAUUUUUGCAUUC-UUGCAGAAAA- ..(((.....((...((((((((..(((.(....))))..))))))))...))...(((..(((((((.((((((((((....)))))....-.)))))...))))))-)))).)))..- ( -24.80) >DroYak_CAF1 12902 116 - 1 ACUUCAAUUACAACGCCUUCACAAAGUAUGUCAACUAUCUUGUGAAGGAAAUGCUCGUAACAGGAUGCAAGCACAGUGAACACUCACGAAAG-AUUAUUGUUACAUUU-UUGAAGAA-A- .((((((...((...(((((((((..((.(....)))..)))))))))...))...(((((((..((....))..((((....)))).....-....)))))))....-))))))..-.- ( -28.80) >consensus ACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAGGAUGCAAGUACAGUGAAAACUCACAAAUAAAUUAAUGU_AUAUUU_UUACAGAA_A_ ...............(((((((((..((.(....)))..)))))))))...((((.(((......))).))))..((((....))))................................. (-17.52 = -16.60 + -0.92)

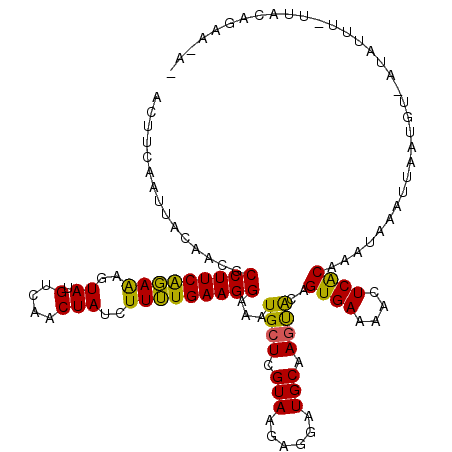
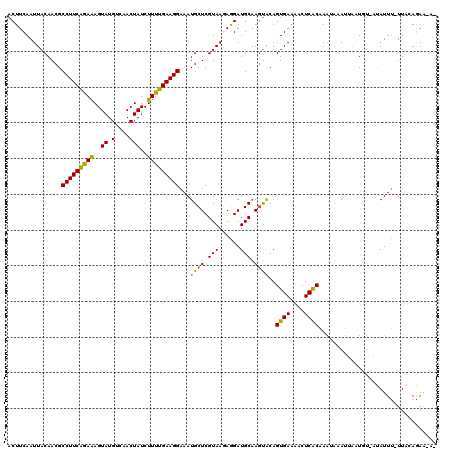
| Location | 17,051,100 – 17,051,220 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051100 120 + 22407834 UUCACUGUACUUGCAUCCUCUUACGAGCAUUUCCUUCAAAAGAUAGUUGACAUACUUUCUGAAGGCGUUGUAAUUGAAGUGUUGCUUAAACAAAAGCUAAAACUUAAAUUGCUGCGAAAU ...........(((.((.......)))))...((((((...((.(((......))).))))))))(((.((((((.((((...((((......))))....)))).)))))).))).... ( -24.60) >DroSec_CAF1 15013 120 + 1 UUCACUGUACUUGCAUCUUCUUACGAGCAUUUCCUUCAAAAGAUAGUUGACAUACUUUCUGAAGGCGUUGUAAUUGAAGUGUUGCUUAAACAAAAGCUAAAACUUAAAUUGCUGCGAAAU ...........(((.((.......)))))...((((((...((.(((......))).))))))))(((.((((((.((((...((((......))))....)))).)))))).))).... ( -25.10) >DroSim_CAF1 9297 120 + 1 UUCACUGUAGUUGCAUCUUCUUACGAGCAUUUCCUUCAAAAGAUAGUUGACAUACUUUCUGAAGGCGUUGUAAUUGAAGUGUUGCUUAAACAAAAGCUAAAACUUAAAUUGCUGCGAAAU ...........(((.((.......)))))...((((((...((.(((......))).))))))))(((.((((((.((((...((((......))))....)))).)))))).))).... ( -25.10) >DroEre_CAF1 14885 120 + 1 UUCACUGCACUGGCAUCCUGUUACGAGCAUUUCCUUCAAAAGAUAGUUGACAUACUCUCCGAAGGCGUUGUAAUUGAAGUGUUGCUUAAACAAAAGCUAAAACUUAAAUUGCUGCGAAAU .....(((.(((((.....)))).).)))...(((((....((.(((......))).)).)))))(((.((((((.((((...((((......))))....)))).)))))).))).... ( -25.00) >DroYak_CAF1 12938 120 + 1 UUCACUGUGCUUGCAUCCUGUUACGAGCAUUUCCUUCACAAGAUAGUUGACAUACUUUGUGAAGGCGUUGUAAUUGAAGUGUUGCUUAAACAAAAGCUAAAACUUAAAUUGCUGCGAAAU ......(((((((..........)))))))..(((((((((...(((......))))))))))))(((.((((((.((((...((((......))))....)))).)))))).))).... ( -33.00) >consensus UUCACUGUACUUGCAUCCUCUUACGAGCAUUUCCUUCAAAAGAUAGUUGACAUACUUUCUGAAGGCGUUGUAAUUGAAGUGUUGCUUAAACAAAAGCUAAAACUUAAAUUGCUGCGAAAU ............((.((.......))))....((((((...((.(((......))).))))))))(((.((((((.((((...((((......))))....)))).)))))).))).... (-22.66 = -23.06 + 0.40)

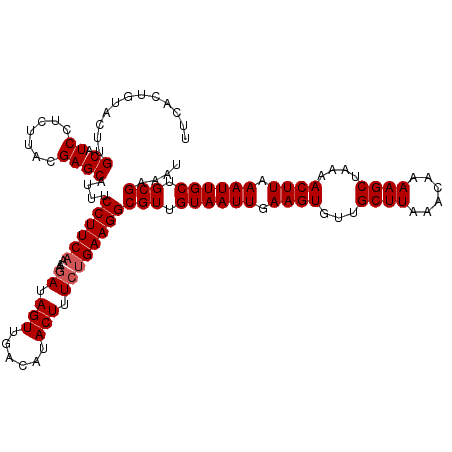
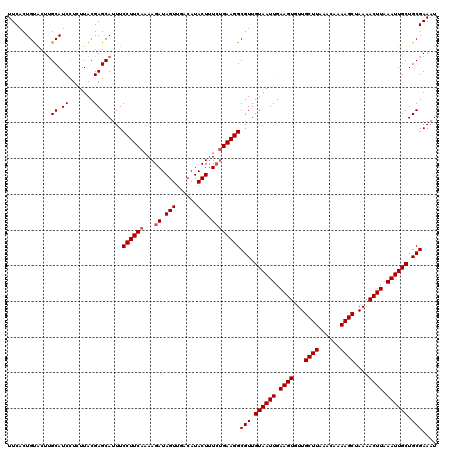
| Location | 17,051,100 – 17,051,220 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -24.62 |
| Energy contribution | -23.98 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17051100 120 - 22407834 AUUUCGCAGCAAUUUAAGUUUUAGCUUUUGUUUAAGCAACACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAGGAUGCAAGUACAGUGAA ..(((((.(.((((.((((.((.((((......)))))).)))).)))).).((.(((((((((..((.(....)))..)))))))))..((.(((....))).))....))...))))) ( -27.00) >DroSec_CAF1 15013 120 - 1 AUUUCGCAGCAAUUUAAGUUUUAGCUUUUGUUUAAGCAACACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAAGAUGCAAGUACAGUGAA ..(((((.(.((((.((((.((.((((......)))))).)))).)))).).((.(((((((((..((.(....)))..)))))))))...(((((.......)).))).))...))))) ( -24.40) >DroSim_CAF1 9297 120 - 1 AUUUCGCAGCAAUUUAAGUUUUAGCUUUUGUUUAAGCAACACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAAGAUGCAACUACAGUGAA ..(((((.((((((.((((.((.((((......)))))).)))).))))......(((((((((..((.(....)))..)))))))))....))((....)).............))))) ( -24.30) >DroEre_CAF1 14885 120 - 1 AUUUCGCAGCAAUUUAAGUUUUAGCUUUUGUUUAAGCAACACUUCAAUUACAACGCCUUCGGAGAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAACAGGAUGCCAGUGCAGUGAA ..(((((.((((((.((((.((.((((......)))))).)))).))))......((((((((..(((.(....))))..))))))))....(((.(((......))).))))).))))) ( -24.00) >DroYak_CAF1 12938 120 - 1 AUUUCGCAGCAAUUUAAGUUUUAGCUUUUGUUUAAGCAACACUUCAAUUACAACGCCUUCACAAAGUAUGUCAACUAUCUUGUGAAGGAAAUGCUCGUAACAGGAUGCAAGCACAGUGAA ..(((((.(.((((.((((.((.((((......)))))).)))).)))).)....(((((((((..((.(....)))..)))))))))...((((.(((......))).))))..))))) ( -28.50) >consensus AUUUCGCAGCAAUUUAAGUUUUAGCUUUUGUUUAAGCAACACUUCAAUUACAACGCCUUCAGAAAGUAUGUCAACUAUCUUUUGAAGGAAAUGCUCGUAAGAGGAUGCAAGUACAGUGAA ..((((((((((((.((((.((.((((......)))))).)))).))))......(((((((((..((.(....)))..)))))))))....))).(((......))).......))))) (-24.62 = -23.98 + -0.64)

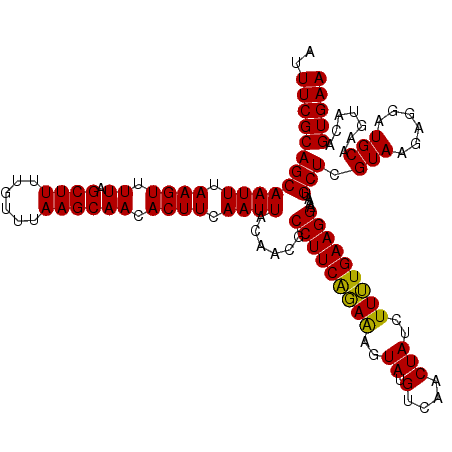
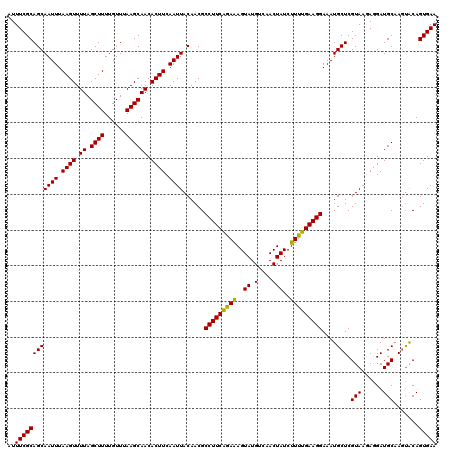
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:16 2006