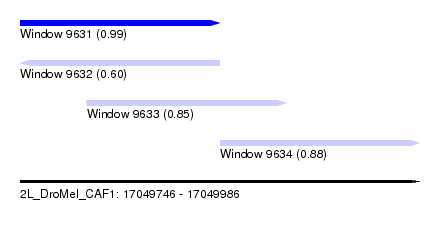
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,049,746 – 17,049,986 |
| Length | 240 |
| Max. P | 0.992879 |

| Location | 17,049,746 – 17,049,866 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049746 120 + 22407834 GUGCACAAAAGUACACAGAAGUUUUAUGAAAGAAAAUGAAAUGAAAAUCCGGUUGCAAUCCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAA ((((......))))......(((((((........))))))).........(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))..... ( -26.30) >DroSec_CAF1 13636 120 + 1 GUGCACAGAAGUACACAGAAGUUUUAUGAAAGAAAAUGAAAUGAAAAUCCGGUUGCAAUCCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAA ((((......))))......(((((((........))))))).........(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))..... ( -26.30) >DroSim_CAF1 7901 120 + 1 GUGCACAAAAGUACACAGAAGUUUUAUGAAAGAAAAUGAAAUGAAAAUCCGGUUGCAAUUCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAA ((((......))))......(((((((........))))))).........(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))..... ( -26.30) >DroEre_CAF1 13590 120 + 1 GUGCACAAAAGUACACAGAAGUUUUAUGAAAGAAAAUGAAAUGAAAAUCCGGUUGCAGUCCCUCAAAGCUUCGUAAACUGAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAA ((((......))))......(((((((........))))))).........(((((.((..((.((((((....((((((((......))))))))..)))))).))...)).))))).. ( -26.70) >DroYak_CAF1 11595 116 + 1 GUGCACAAAAGUACACAGAAGUUUUAUGAAAGAAAAUGAAAUGAAAAUCCGGUUGCAGUCCCUCAAAGCU----AAACCAAGGAAAUCUUUAGUUUUUGGCUUUAAGCUCGCGGCAACAA ((((......))))......(((((((........))))))).........(((((.((..((.((((((----((((.((((....)))).))..)))))))).))...)).))))).. ( -24.90) >consensus GUGCACAAAAGUACACAGAAGUUUUAUGAAAGAAAAUGAAAUGAAAAUCCGGUUGCAAUCCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAA ((((......))))......(((((((........))))))).........(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))..... (-25.10 = -25.10 + -0.00)

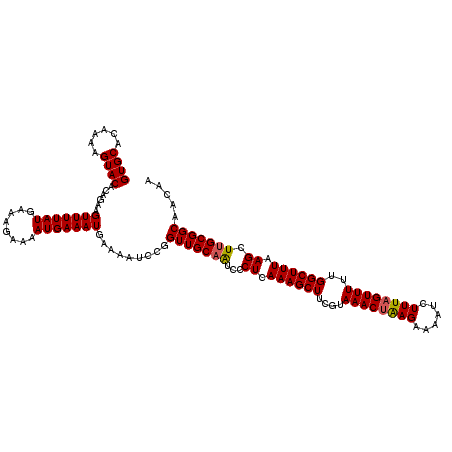

| Location | 17,049,746 – 17,049,866 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.04 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049746 120 - 22407834 UUGUUGCCGCAAGCUUAAAGCCAAAAACUAAAGAUUUUCUUAGUUUACGAAGCUUUGAGGGAUUGCAACCGGAUUUUCAUUUCAUUUUCUUUCAUAAAACUUCUGUGUACUUUUGUGCAC ..((((((....)((((((((...(((((((........))))))).....)))))))).....)))))...................................((((((....)))))) ( -24.70) >DroSec_CAF1 13636 120 - 1 UUGUUGCCGCAAGCUUAAAGCCAAAAACUAAAGAUUUUCUUAGUUUACGAAGCUUUGAGGGAUUGCAACCGGAUUUUCAUUUCAUUUUCUUUCAUAAAACUUCUGUGUACUUCUGUGCAC ..((((((....)((((((((...(((((((........))))))).....)))))))).....)))))...................................((((((....)))))) ( -24.70) >DroSim_CAF1 7901 120 - 1 UUGUUGCCGCAAGCUUAAAGCCAAAAACUAAAGAUUUUCUUAGUUUACGAAGCUUUGAGGAAUUGCAACCGGAUUUUCAUUUCAUUUUCUUUCAUAAAACUUCUGUGUACUUUUGUGCAC ..((((((....)((((((((...(((((((........))))))).....)))))))).....)))))...................................((((((....)))))) ( -24.70) >DroEre_CAF1 13590 120 - 1 UUGUUGCCGCAAGCUUAAAGCCAAAAACUAAAGAUUUUCUCAGUUUACGAAGCUUUGAGGGACUGCAACCGGAUUUUCAUUUCAUUUUCUUUCAUAAAACUUCUGUGUACUUUUGUGCAC ..((((((....)((((((((...(((((..((.....)).))))).....)))))))).....)))))...................................((((((....)))))) ( -22.40) >DroYak_CAF1 11595 116 - 1 UUGUUGCCGCGAGCUUAAAGCCAAAAACUAAAGAUUUCCUUGGUUU----AGCUUUGAGGGACUGCAACCGGAUUUUCAUUUCAUUUUCUUUCAUAAAACUUCUGUGUACUUUUGUGCAC ..((((((....)((((((((...(((((((.(....).)))))))----.)))))))).....)))))...................................((((((....)))))) ( -25.00) >consensus UUGUUGCCGCAAGCUUAAAGCCAAAAACUAAAGAUUUUCUUAGUUUACGAAGCUUUGAGGGAUUGCAACCGGAUUUUCAUUUCAUUUUCUUUCAUAAAACUUCUGUGUACUUUUGUGCAC ..((((((....)((((((((...(((((((........))))))).....)))))))).....)))))...................................((((((....)))))) (-24.20 = -24.24 + 0.04)

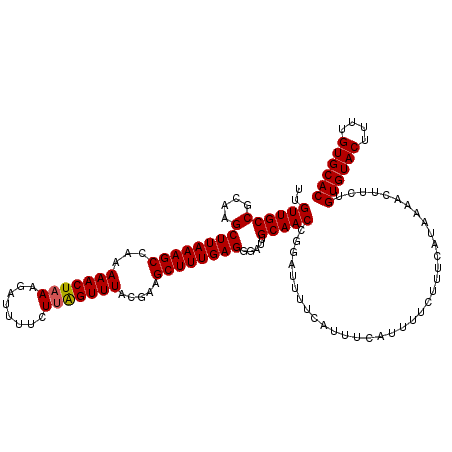
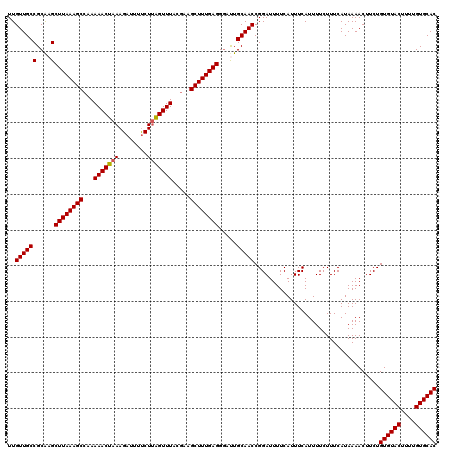
| Location | 17,049,786 – 17,049,906 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -24.00 |
| Energy contribution | -25.50 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049786 120 + 22407834 AUGAAAAUCCGGUUGCAAUCCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAAUUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGC ........((.(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))...........((((((...((....))........)))))))). ( -26.70) >DroSec_CAF1 13676 120 + 1 AUGAAAAUCCGGUUGCAAUCCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAAUUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGC ........((.(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))...........((((((...((....))........)))))))). ( -26.70) >DroSim_CAF1 7941 120 + 1 AUGAAAAUCCGGUUGCAAUUCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAAUUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGC ........((.(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))...........((((((...((....))........)))))))). ( -26.70) >DroEre_CAF1 13630 120 + 1 AUGAAAAUCCGGUUGCAGUCCCUCAAAGCUUCGUAAACUGAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAAUUAAAUGGGAAUAAGCUGAAAAGAAAAUGAGAUUUCCGGC ........((.(((((.((..((.((((((....((((((((......))))))))..)))))).))...)).)))))........((((((...((....))........)))))))). ( -27.10) >DroYak_CAF1 11635 116 + 1 AUGAAAAUCCGGUUGCAGUCCCUCAAAGCU----AAACCAAGGAAAUCUUUAGUUUUUGGCUUUAAGCUCGCGGCAACAAUUAAAUGGGAAUAAGCUGAAAAGAAAAUAAGAUUUCCGGC ........((((((..(((........)))----.))))..(((((((((...((((..((((....((((.(....).......))))...))))..))))......))))))))))). ( -27.30) >consensus AUGAAAAUCCGGUUGCAAUCCCUCAAAGCUUCGUAAACUAAGAAAAUCUUUAGUUUUUGGCUUUAAGCUUGCGGCAACAAUUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGC ........((.(((((((...((.((((((....((((((((......))))))))..)))))).)).)))))))...........((((((...((....))........)))))))). (-24.00 = -25.50 + 1.50)

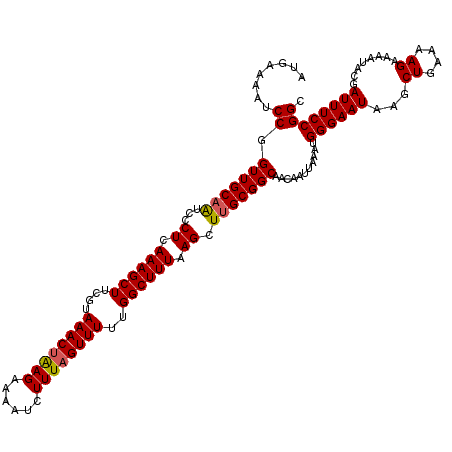
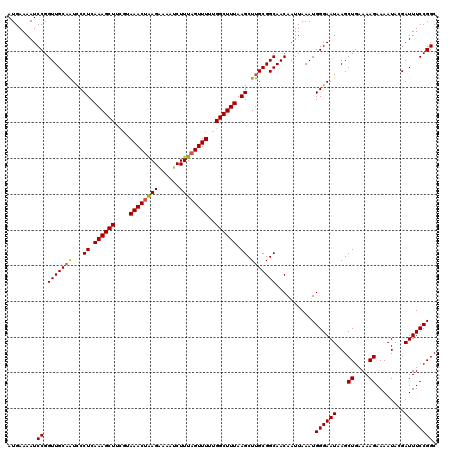
| Location | 17,049,866 – 17,049,986 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.59 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049866 120 + 22407834 UUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGCUUGGAAUCCUUUGACAAAACACUUUCUGGAAGCACAGAAAACAGGGAAAAUCAAUUUGGAACUGUGAGGGAUUUUCCACU .......(((.(((((((....((((......)))))))))))(((((((((..........((((((......))))))((((....((.....))....))))))))))))))))... ( -27.40) >DroSec_CAF1 13756 120 + 1 UUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGCUUGGAAUCCUUUGACAAAACACUUUCUGGAAGCACAGAAAACAGGGAAAAUCAAUUUGGAACUGUGAGGGAUUUUCCACU .......(((.(((((((....((((......)))))))))))(((((((((..........((((((......))))))((((....((.....))....))))))))))))))))... ( -27.40) >DroSim_CAF1 8021 120 + 1 UUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGCUUGGAAUCCUUUGACAAAACACUUUCUGGAAGCACAGAAAACAGGGAAAAUCAAUUUGGAACUGUGAGGGAUUUUCCACU .......(((.(((((((....((((......)))))))))))(((((((((..........((((((......))))))((((....((.....))....))))))))))))))))... ( -27.40) >DroEre_CAF1 13710 120 + 1 UUAAAUGGGAAUAAGCUGAAAAGAAAAUGAGAUUUCCGGCUUGGAAUCCUUUGACAAAACACUUUCUGGAAUAACAGAAAACAGGGACAAUCAAUUUGGGACUGUAGGGGAUUUUCCACU .......(((.(((((((....((((......)))))))))))(((((((((..........((((((......))))))((((...(((.....)))...))))))))))))))))... ( -29.70) >DroYak_CAF1 11711 120 + 1 UUAAAUGGGAAUAAGCUGAAAAGAAAAUAAGAUUUCCGGCUGGGAAUGCUUUGACAAAACACUUUCUGGAAUAACAGAAAACAGGGACAAUCAAUUUGGGACAGCAAGGGAUUUUCCACU .....(((((((...((....))........)))))))..(((((((.(((((.(.......((((((......))))))...(...(((.....)))...).)))))).).)))))).. ( -22.40) >consensus UUAAAUGGGAAUAAGCUGAAAAGAAAAUACGAUUUCCGGCUUGGAAUCCUUUGACAAAACACUUUCUGGAAGCACAGAAAACAGGGAAAAUCAAUUUGGAACUGUGAGGGAUUUUCCACU .......(((.(((((((....((((......)))))))))))((((((((((.........((((((......)))))).(((.................))))))))))))))))... (-23.55 = -23.59 + 0.04)

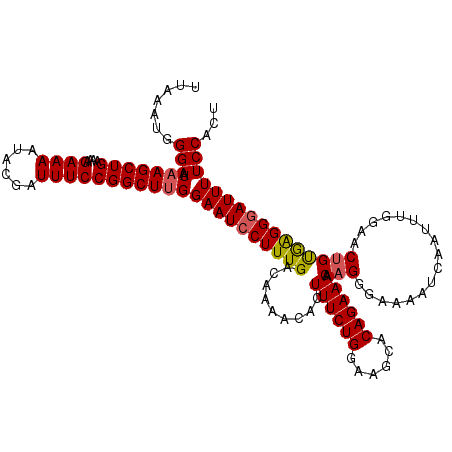
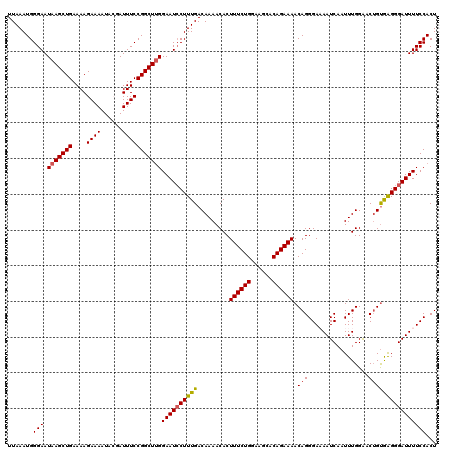
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:12 2006