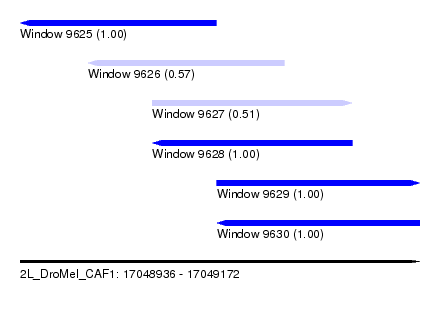
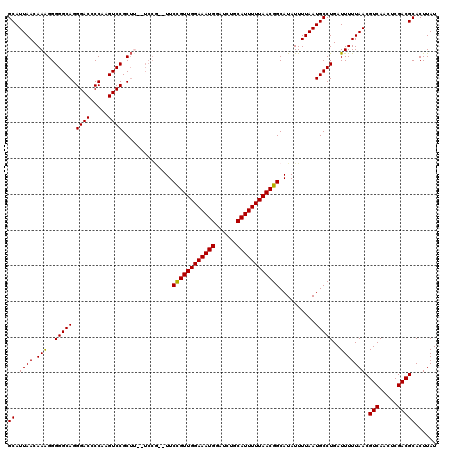
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,048,936 – 17,049,172 |
| Length | 236 |
| Max. P | 0.999830 |

| Location | 17,048,936 – 17,049,052 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.40 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -31.58 |
| Energy contribution | -31.26 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17048936 116 - 22407834 GCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUU--UCCG--UUCCGUUGGAAAUGUAUCUGCAUUUUUAACGGCAUAUUUUAAUGCCUGAUUUUUAACGUCAACUCGACGCACUUAU (((((((.((((.((((.((....))..)))).)))--)..(--(.(((((((((((((....))))))))))))).))...)))))))...........((((.....))))....... ( -35.70) >DroSec_CAF1 12996 116 - 1 GCAUUAACAAAGGGGGCAGGGACACCAAGUCCGCUU--UCCG--UUCUGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUAUUUUAAUGCCUGAUUUUUAACGUCAACUCGACGCACUUAU (((((((....(..(((..((((.....))))))).--.).(--(.((((((((((((......)))))))))))).))...)))))))...........((((.....))))....... ( -32.10) >DroSim_CAF1 7089 116 - 1 GCAUUAACAAAGGGGGCAGGGACACCAAGUCCGCUU--UCCG--UUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUAUUUUAAUGCCUGAUUUUUAACGUCAACUCGACGCACUUAU (((((((....(..(((..((((.....))))))).--.).(--(.((((((((((((......)))))))))))).))...)))))))...........((((.....))))....... ( -34.30) >DroEre_CAF1 12751 111 - 1 GCAUUAACAAGGGGGGCAGGGACCCCAAGUCCGC---------UUUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUAUUUUAAUGCCUGAUUUUUAACGUCAACUCGACGCACUUAU ((........((..(((..((((.....))))))---------)..))((((((((((......))))))))))(((((......)))))...........(((.....)))))...... ( -34.30) >DroYak_CAF1 10752 120 - 1 GCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUGGAUCCAUUUUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUAUUUUAAUGCCUGAUUUUUAACGUCAACUCGACGCACUUAU ((.........((((.......))))(((((.((.((((((((((((....)))))))))))))).........(((((......))))).))))).....(((.....)))))...... ( -38.10) >consensus GCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUU__UCCG__UUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUAUUUUAAUGCCUGAUUUUUAACGUCAACUCGACGCACUUAU ((.((((.(((..(((((.((((.....))))..............((((((((((((......))))))))))))..........)))))..))))))).(((.....)))))...... (-31.58 = -31.26 + -0.32)

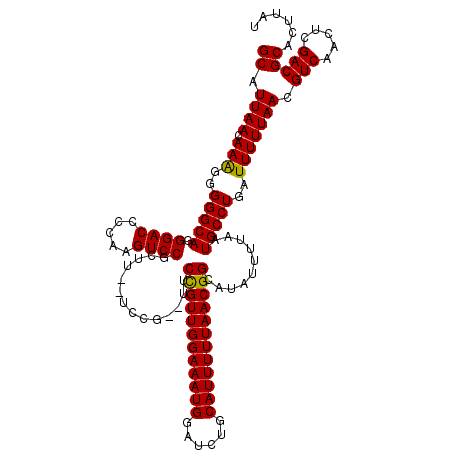
| Location | 17,048,976 – 17,049,092 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17048976 116 - 22407834 CAAGUGGCACCGCAUCACUAGCAGCUGUGGCUUAUGAAGUGCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUU--UCCG--UUCCGUUGGAAAUGUAUCUGCAUUUUUAACGGCAUA .((((((.((.((((((..(((.......)))..)))..))).........((((.......))))..))))))))--...(--(.(((((((((((((....))))))))))))).)). ( -37.90) >DroSec_CAF1 13036 116 - 1 CAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAAGGGGGCAGGGACACCAAGUCCGCUU--UCCG--UUCUGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUA ....(((((.........)))))(((((.......((((((((......(((.((((.((....))..)))).)))--((((--(((.....).))))))..))))))))..)))))... ( -33.20) >DroSim_CAF1 7129 116 - 1 CAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAAGGGGGCAGGGACACCAAGUCCGCUU--UCCG--UUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUA ...(((....))).(((..((((....))))...))).((((.........(..(((..((((.....))))))).--.)..--...(((((((((((......))))))))))))))). ( -35.40) >DroEre_CAF1 12791 111 - 1 CAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAGGGGGGCAGGGACCCCAAGUCCGC---------UUUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUA .((((((.((.((((((..((((....))))...)))..)))........((((........))))..))))))---------)).((((((((((((......)))))))))))).... ( -38.30) >DroYak_CAF1 10792 116 - 1 CAAGUGGCACCGCAUCACUGGCAGCUGUGGCUUA----AUGCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUGGAUCCAUUUUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUA ...(((....)))..........(((((.....(----((((........((.((((.((....))..)))).))((((((((((((....)))))))))))))))))....)))))... ( -40.70) >consensus CAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUU__UCCG__UUCCGUUGGAAAUGGAUCUGCAUUUUUAACGGCAUA ..(((((.((.((((.......(((....)))......)))).........((((.......))))..)))))))...........((((((((((((......)))))))))))).... (-30.00 = -30.28 + 0.28)

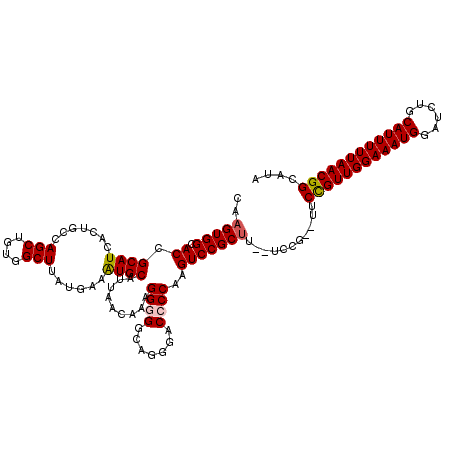
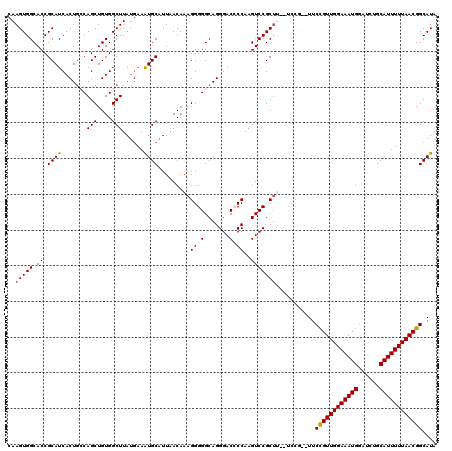
| Location | 17,049,014 – 17,049,132 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -29.26 |
| Energy contribution | -30.90 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049014 118 + 22407834 GA--AAGCGGACUUGGGGUCCCUGCCCCCUUUGUUAAUGCACUUCAUAAGCCACAGCUGCUAGUGAUGCGGUGCCACUUGCAUAUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUU ..--..((((....(((((....)))))....(((((.(((((((((.(((.......))).))))...)))))(((((((.....)))))))...))))).)))).............. ( -34.60) >DroSec_CAF1 13074 118 + 1 GA--AAGCGGACUUGGUGUCCCUGCCCCCUUUGUUAAUGCAUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUU ..--.((.((((.....)))))).....((((((....(((((((((..(((((((((((((.((...)).)))))...)).))))))..))))....)))))...))))))........ ( -37.50) >DroSim_CAF1 7167 118 + 1 GA--AAGCGGACUUGGUGUCCCUGCCCCCUUUGUUAAUGCAUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUU ..--.((.((((.....)))))).....((((((....(((((((((..(((((((((((((.((...)).)))))...)).))))))..))))....)))))...))))))........ ( -37.50) >DroEre_CAF1 12828 114 + 1 ------GCGGACUUGGGGUCCCUGCCCCCCUUGUUAAUGCAUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCGAACUUU ------((((....(((((....)))))))((((....(((((((((..(((((((((((((.((...)).)))))...)).))))))..))))....)))))...)))).))....... ( -38.20) >DroYak_CAF1 10832 116 + 1 GAUCCAGCGGACUUGGGGUCCCUGCCCCCUUUGUUAAUGCAU----UAAGCCACAGCUGCCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUU ....(((.((((.....)))))))....((((((....((((----((((.(((.(((((.(....))))))(((((......)))))..)))..))))))))...))))))........ ( -38.00) >consensus GA__AAGCGGACUUGGGGUCCCUGCCCCCUUUGUUAAUGCAUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUU ......((((((.....))))..))...((((((....(((((((((..(((((((((((((.((...)).)))))...)).))))))..)))))....))))...))))))........ (-29.26 = -30.90 + 1.64)

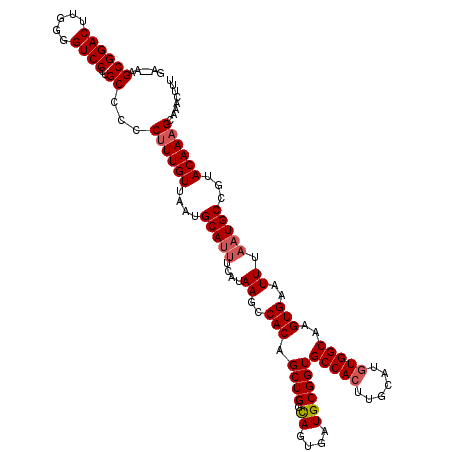
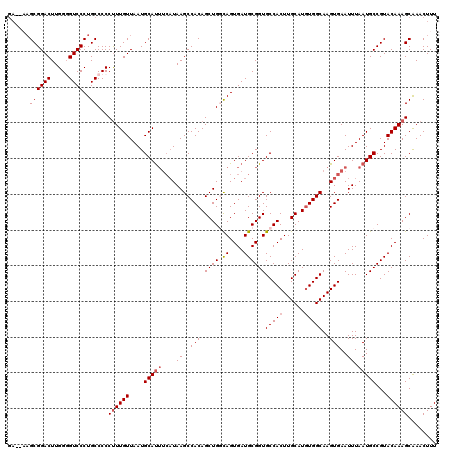
| Location | 17,049,014 – 17,049,132 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -35.48 |
| Energy contribution | -35.20 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049014 118 - 22407834 AAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCAUAUGCAAGUGGCACCGCAUCACUAGCAGCUGUGGCUUAUGAAGUGCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUU--UC ...((((.((((((...((((....((((...(((((((((.(((((.......))))).)))..))))))...))))))))....)))))).))))..((((.....))))....--.. ( -36.90) >DroSec_CAF1 13074 118 - 1 AAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAAGGGGGCAGGGACACCAAGUCCGCUU--UC ...((((.((((((...(((((....(((...((((((.((...(((((.........)))))))))))))...))))))))....)))))).))))..((((.....))))....--.. ( -39.00) >DroSim_CAF1 7167 118 - 1 AAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAAGGGGGCAGGGACACCAAGUCCGCUU--UC ...((((.((((((...(((((....(((...((((((.((...(((((.........)))))))))))))...))))))))....)))))).))))..((((.....))))....--.. ( -39.00) >DroEre_CAF1 12828 114 - 1 AAAGUUCGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAGGGGGGCAGGGACCCCAAGUCCGC------ ...((((.((((((...(((((....(((...((((((.((...(((((.........)))))))))))))...))))))))....)))))).))))..((((.....))))..------ ( -39.90) >DroYak_CAF1 10832 116 - 1 AAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGGCAGCUGUGGCUUA----AUGCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUGGAUC ...((((.((((((...(((((((........(((((((((.(((((.......))))).)))..)))))))))----))))....)))))).))))((((((.....)))).))..... ( -38.00) >consensus AAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAUGCAUUAACAAAGGGGGCAGGGACCCCAAGUCCGCUU__UC ...((((.((((((...(((((....((((((((.....))))))))..(((((.(.......).))))).......)))))....)))))).))))..((((.....))))........ (-35.48 = -35.20 + -0.28)

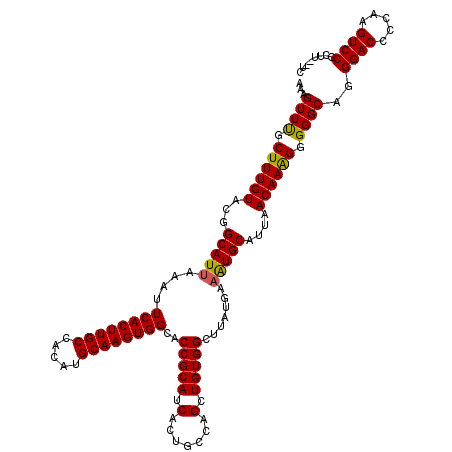

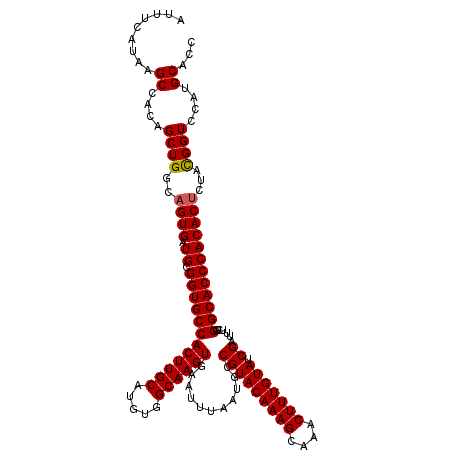
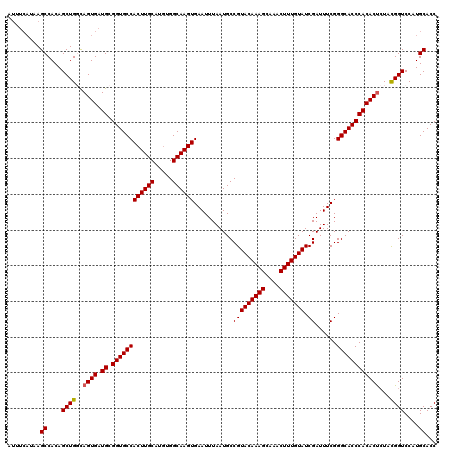
| Location | 17,049,052 – 17,049,172 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -36.98 |
| Energy contribution | -37.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049052 120 + 22407834 ACUUCAUAAGCCACAGCUGCUAGUGAUGCGGUGCCACUUGCAUAUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUUGUAUCGAUUUCGGGCACCCACACUAUAUGGUCCAUGCACC ...((((((((....)))..(((((.((.((((((((((((.....))))))...........(((((((((....))))))).))......)))))))))))))))))).......... ( -37.20) >DroSec_CAF1 13112 120 + 1 AUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUUGUAUCGAUUUCGGGCACCCACACUCUACGGUCCAUGCACC .........((....((((..((((.((.((((((((((((.....))))))...........(((((((((....))))))).))......))))))))))))...))))....))... ( -38.40) >DroSim_CAF1 7205 120 + 1 AUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUUGUAUCGAUUUCGGGCACCCACACUCUACGGUCCAUGCACC .........((....((((..((((.((.((((((((((((.....))))))...........(((((((((....))))))).))......))))))))))))...))))....))... ( -38.40) >DroEre_CAF1 12862 120 + 1 AUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCGAACUUUGUAUCGAUUUCGGGCACCCACACCCUACGGUCCAUGCACC ...((((..((((....)))).))))...((((((((((((.....)))))))........(((((((((((....)))))).........(((........))).)))))....))))) ( -37.60) >DroYak_CAF1 10872 116 + 1 AU----UAAGCCACAGCUGCCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUUGUAUCGAUUUCGGGCACCCACACUCUACGGUCCAUGCACC ..----...((....((((..((((.((.((((((((((((.....))))))...........(((((((((....))))))).))......))))))))))))...))))....))... ( -37.30) >consensus AUUUCAUAAGCCACAGCUGGCAGUGAUGCGGUGCCACUUGCAUGUGGCAAGUGAAUUUAAUGCCGUACAAAGCAAACUUUGUAUCGAUUUCGGGCACCCACACUCUACGGUCCAUGCACC .........((....((((..((((.((.((((((((((((.....))))))...........(((((((((....))))))).))......))))))))))))...))))....))... (-36.98 = -37.02 + 0.04)

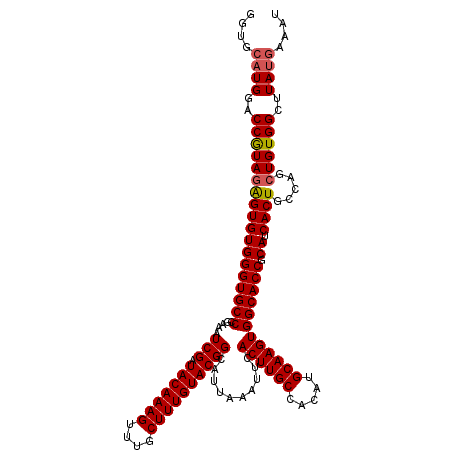
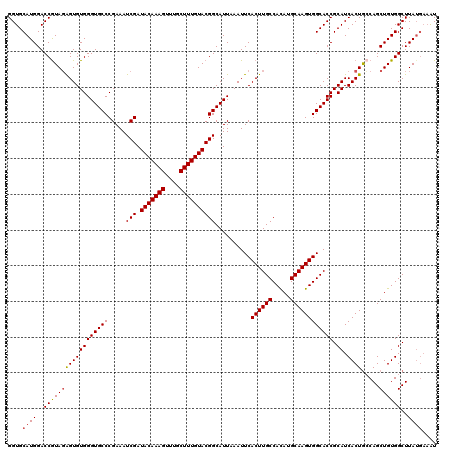
| Location | 17,049,052 – 17,049,172 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -37.98 |
| Energy contribution | -38.26 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17049052 120 - 22407834 GGUGCAUGGACCAUAUAGUGUGGGUGCCCGAAAUCGAUACAAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCAUAUGCAAGUGGCACCGCAUCACUAGCAGCUGUGGCUUAUGAAGU ....((((..((((((((((((((((((.....(((.(((((((....))))))))))..........((((((.....)))))))))))).)).))))).....)))))..)))).... ( -41.20) >DroSec_CAF1 13112 120 - 1 GGUGCAUGGACCGUAGAGUGUGGGUGCCCGAAAUCGAUACAAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAU (((((..........((((...((((((((....)).(((((((....))))))).))))))..))))((((((.....)))))).)))))...(((..((((....))))...)))... ( -41.10) >DroSim_CAF1 7205 120 - 1 GGUGCAUGGACCGUAGAGUGUGGGUGCCCGAAAUCGAUACAAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAU (((((..........((((...((((((((....)).(((((((....))))))).))))))..))))((((((.....)))))).)))))...(((..((((....))))...)))... ( -41.10) >DroEre_CAF1 12862 120 - 1 GGUGCAUGGACCGUAGGGUGUGGGUGCCCGAAAUCGAUACAAAGUUCGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAU (((.(((((...((((((((((((((((((....)).(((((((....))))))).))))).....((((((((.....))))))))..))))))).))))...))))))))........ ( -43.90) >DroYak_CAF1 10872 116 - 1 GGUGCAUGGACCGUAGAGUGUGGGUGCCCGAAAUCGAUACAAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGGCAGCUGUGGCUUA----AU (((......)))...(((((..((((((((....)).(((((((....))))))).)))))).....)))))(((((((((.(((((.......))))).)))..))))))...----.. ( -39.00) >consensus GGUGCAUGGACCGUAGAGUGUGGGUGCCCGAAAUCGAUACAAAGUUUGCUUUGUACGGCAUUAAAUUCACUUGCCACAUGCAAGUGGCACCGCAUCACUGCCAGCUGUGGCUUAUGAAAU ....((((..((((((((((((((((((.....(((.(((((((....))))))))))..........((((((.....)))))))))))).)).)))).....))))))..)))).... (-37.98 = -38.26 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:08 2006