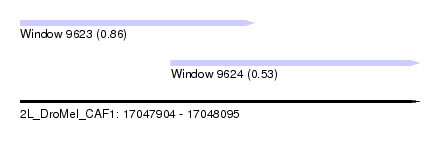
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,047,904 – 17,048,095 |
| Length | 191 |
| Max. P | 0.863439 |

| Location | 17,047,904 – 17,048,016 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -32.56 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
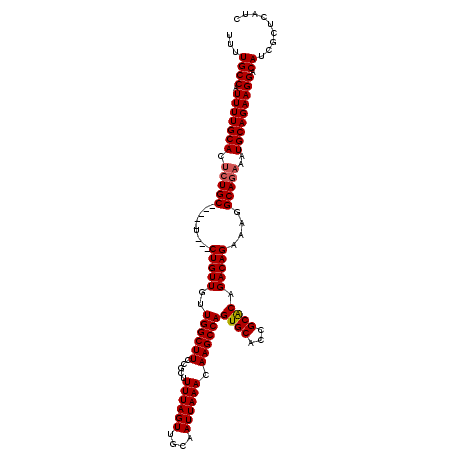
>2L_DroMel_CAF1 17047904 112 + 22407834 UUUUGCCAUUUUGCACUCUGC-----U---CUGUUGUUGGCUUCCGCUUUUAGUUGCAAUUAAACAAGCCAGUGCACCGCACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUC ...((((.(((((((.(((((-----(---(((((..((((((.....((((((....)))))).))))))((((...)))).))))))...)))))..))))))))).))......... ( -33.10) >DroSec_CAF1 12012 112 + 1 UUUUGCCAUUUUGCACUCUGC-----U---CUGUUGCUGGCUUCCGCUUUUAGUUGCAAUUAAACAAGCCAGUGCACCGCACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUC ...((((.(((((((.(((((-----(---(((((((((((((.....((((((....)))))).))))))))((...))...))))))...)))))..))))))))).))......... ( -33.10) >DroSim_CAF1 5659 112 + 1 UUUUGCCAUUUUGCACUCUGC-----U---CUGUUGUUGGCUUCCGCUUUUAGUUGCAAUUAAACAAGCCAGUGCACCGCACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUC ...((((.(((((((.(((((-----(---(((((..((((((.....((((((....)))))).))))))((((...)))).))))))...)))))..))))))))).))......... ( -33.10) >DroEre_CAF1 11732 112 + 1 UUUUGCCAUUUUGCAGCGUGC-----U---CUGUUGUUGGCUUCCGCUUUUAGUUGCAAUUAAACAAGCCAGUGCACCGCGCAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUC ...((((.(((((((.(.(((-----(---(((((..((((((.....((((((....)))))).))))))((((...)))).))))))...))))...))))))))).))......... ( -29.10) >DroYak_CAF1 9683 120 + 1 UUUUGCCAUUUUGCACUCUGCAUUGCUCUGCUGUUGUUGGCUUCCGCUUUUAGUUGCAAUUAAACAAGCCAGUGCACCGCACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUC ....((.....((..(((((((((..((((((.((((((((((.....((((((....)))))).))))).((((...)))).)))))...))))))))))))).))..))..))..... ( -34.40) >consensus UUUUGCCAUUUUGCACUCUGC_____U___CUGUUGUUGGCUUCCGCUUUUAGUUGCAAUUAAACAAGCCAGUGCACCGCACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUC ...((((.(((((((.(((((.........(((((..((((((.....((((((....)))))).))))))((((...)))).)))))....)))))..))))))))).))......... (-27.98 = -28.22 + 0.24)

| Location | 17,047,976 – 17,048,095 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.35 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -26.98 |
| Energy contribution | -26.82 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
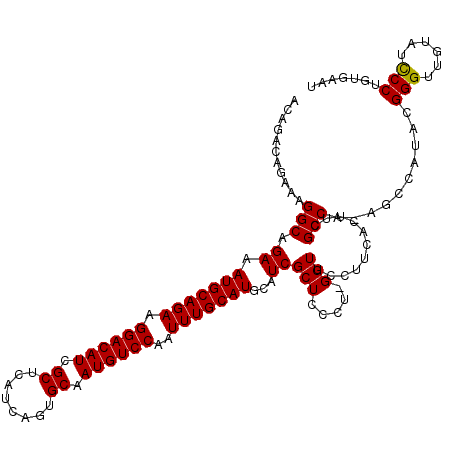
>2L_DroMel_CAF1 17047976 119 + 22407834 ACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUCAGUGCAAUGUCCAAUUUGCAUGCAUCGCUCCCU-GGUUCCUUCACUUCGCCAAAGCCAUACGGGUUGUAUCCCUCUGAAU .....((((..(((.((.(((((((.((((((.((........)).))))))..)))))))...))......(-(((...........))))..)))....(((......)))))))... ( -34.40) >DroSec_CAF1 12084 119 + 1 ACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUCAGUGCAAUGUCCAAUUUGCAUGCAUCGCUCCCU-GGUUCCUUCACUUCGCCAUAGCCAUACGGGUUGUAUCCCUCUGAAU .....((((..(((.((.(((((((.((((((.((........)).))))))..)))))))...))......(-(((...........))))..)))....(((......)))))))... ( -34.20) >DroSim_CAF1 5731 119 + 1 ACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUCAGUGCAAUGUCCAAUUUGCAUGCAUCGCUCCCU-GGUUCCUUCACUUCGCCAUAGCCAUACGGGUUGAAUCCCUGUGAAU .....(((...(((.((.(((((((.((((((.((........)).))))))..)))))))...)))))..))-).....(((((...((....)).....(((......))).))))). ( -32.20) >DroEre_CAF1 11804 113 + 1 GCAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUCAGUGCAAUGUCCAAUUUGCAUGCAUCGCUCCCU-GGUUCCUUCACUUCGCC------ACACGGGUUGUAUCCCUGUGAAU .....(((...(((.((.(((((((.((((((.((........)).))))))..)))))))...)))))..))-)...............(------(((.(((......)))))))... ( -32.50) >DroYak_CAF1 9763 114 + 1 ACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUCAGUGCAAUGUCCAAUUUGCAUGCAUCGCUCCCCGGGUUCCUUCACUUCGCC------AAACGGGUUCUAUUCCUGUGCAU ((((..((((.(((.((.(((((((.((((((.((........)).))))))..)))))))...))))).(((((((...........)))------...))))))))....)))).... ( -30.80) >consensus ACAGACAGAAAGGCAGAAAUGCAGAAGGACAUCGCUCAUCAGUGCAAUGUCCAAUUUGCAUGCAUCGCUCCCU_GGUUCCUUCACUUCGCCA_AGCCAUACGGGUUGUAUCCCUGUGAAU ...........(((.((.(((((((.((((((.((........)).))))))..)))))))...))(((.....)))...........)))..........(((......)))....... (-26.98 = -26.82 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:02 2006