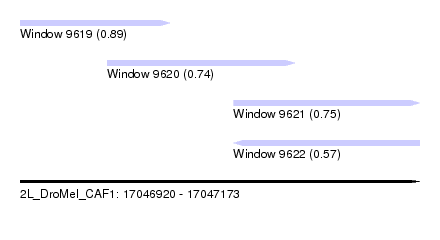
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,046,920 – 17,047,173 |
| Length | 253 |
| Max. P | 0.887678 |

| Location | 17,046,920 – 17,047,015 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.38 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -6.52 |
| Energy contribution | -6.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17046920 95 + 22407834 --AUCACUUUAAAGG----------CUAUUAAAAUGAAG-------------AUGGCUUUAAAAGAUUAAAACUCCUGCUGUUAGGGUAUAGGAAGAGAAAACAGCGACUCAAACAGUUG --....((((.((((----------(((((........)-------------)))))))).))))............((((((.((((....(.........)....)))).)))))).. ( -19.40) >DroSec_CAF1 11019 95 + 1 --AUCACUUUAAAGU----------CUAAAAUAAUUUAG-------------AUGGCUUUAAAAGAUUGAAGCACCAGCUGUUAGGGUGUAGGAAGAGAAAACAGCGACUCAAACAGUUG --...........((----------(((((....)))))-------------)).(((((((....)))))))..((((((((.((((((.(..........).)).)))).)))))))) ( -23.30) >DroSim_CAF1 4675 95 + 1 --AUCACUUUAAAGC----------CUAAAUUUAUUUAG-------------AUGGCUUUAAAAGAUUGAAGCACCCGCUCUUAGGGUGUAGGAAGAGAAAACAGCGACUCAAACAGUUG --.....((((((((----------(.............-------------..))))))))).(((((..((((((.......)))))).....(((..........)))...))))). ( -23.26) >DroEre_CAF1 10729 115 + 1 GUAUAACUUUUAUGCAGUAU-----CUGUUCAAGUGCAGAUACUUACAACUUCCGGCUUCAAAAGGUGGAGACGCCUGCUGUUAGGGUAUAGGAAGAGAAAACAGCGCCUCAAACAGUUG (((((.....)))))(((((-----((((......))))))))).....(((((.(((..((.(((((....)))))....))..)))...)))))......((((..........)))) ( -36.00) >DroYak_CAF1 8634 115 + 1 -----ACUUUUAUACAGUAUUUUAUUUUUUUAAGUAUAGAGACUUCAGUCUUCUGACUUCAAAAGGUGGAGACGCCCGCUGUUAGGGUAUGGGGAGAGAAAACAGCGACUCAAACAGUUG -----.(((((...((((.(((((((((((.((((.(((((((....))).)))))))).)))))))))))))((((.......)))).))..))))).......(((((.....))))) ( -27.50) >consensus __AUCACUUUAAAGC__________CUAUUAAAAUGUAG_____________AUGGCUUUAAAAGAUUGAAACACCCGCUGUUAGGGUAUAGGAAGAGAAAACAGCGACUCAAACAGUUG ......((((...............((((......))))................(((((((.((.(((......))))).)))))))....))))......((((..........)))) ( -6.52 = -6.52 + 0.00)

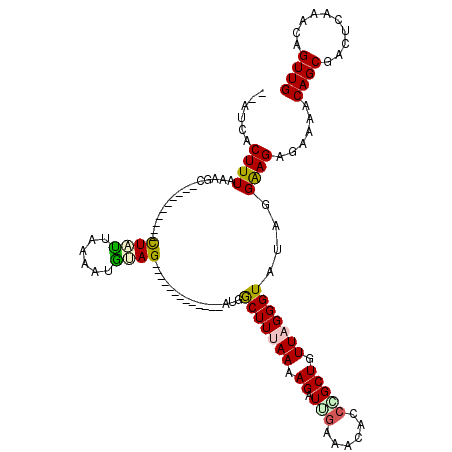

| Location | 17,046,975 – 17,047,094 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.46 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17046975 119 + 22407834 GUUAGGGUAUAGGAAGAGAAAACAGCGACUCAAACAGUUGCAAAUGUUGCAUUUGCAUGCAUGGCCAGUCGGCAGCAAAUUUCACCUGUCCCCUUGCAC-UCCUCUUCACACCCCAAACC (((.(((.....((((((......((((((.....))))))..(((((((....))).)))).((.((..(((((..........)))))..)).))..-..))))))....))).))). ( -33.60) >DroSec_CAF1 11074 120 + 1 GUUAGGGUGUAGGAAGAGAAAACAGCGACUCAAACAGUUGAAAAUGUUGCAUUUGCAUGCAUGGCCAGUCGGCAGCAAAUUUCACCUGUCCCCUUGCACAUCCUCGUUCCACUCCAACCC ((.((((..((((..((((.....(((((((((....))))....))))).(((((.(((.(((....))))))))))))))).))))..)))).))....................... ( -30.90) >DroSim_CAF1 4730 120 + 1 CUUAGGGUGUAGGAAGAGAAAACAGCGACUCAAACAGUUGCAAAUGUUGCAUUUGCAUGCAUGGCCAGUCGGCAGCAAAUUUCACCUGUCCCCUUGCACAUCCUCUUUCCACUCCAACCC .....((.((.(((((((......((((((.....))))))..(((((((....))).)))).((.((..(((((..........)))))..)).)).....))))))).)).))..... ( -32.80) >DroEre_CAF1 10804 115 + 1 GUUAGGGUAUAGGAAGAGAAAACAGCGCCUCAAACAGUUGCAAAUGUUGCAUUUGCAUGCAUGGCCAGUCGGCAGCAAAUUUCACCUGUCCCCUUGCACCUCCUCUACC-----CCUCCC ....(((((.((((.(((..........)))..((((.((((((((...))))))))(((...(((....))).)))........))))...........)))).))))-----)..... ( -31.30) >DroYak_CAF1 8709 115 + 1 GUUAGGGUAUGGGGAGAGAAAACAGCGACUCAAACAGUUGCAAAUGUUGCAUUUGCAUGCAUGGCCAGUCGGCAGCAAAUUUCACCUGUCCCCUCGUACCUCCUCCACC-----CAUGCC .....((((((((..(((......((((.....((((.((((((((...))))))))(((...(((....))).)))........))))....)))).....)))..))-----)))))) ( -38.00) >consensus GUUAGGGUAUAGGAAGAGAAAACAGCGACUCAAACAGUUGCAAAUGUUGCAUUUGCAUGCAUGGCCAGUCGGCAGCAAAUUUCACCUGUCCCCUUGCACAUCCUCUUCCCAC_CCAACCC ((.((((.(((((..((((.((((((((((.....))))))...))))...(((((.(((.(((....))))))))))))))).))))).)))).))....................... (-26.80 = -26.88 + 0.08)

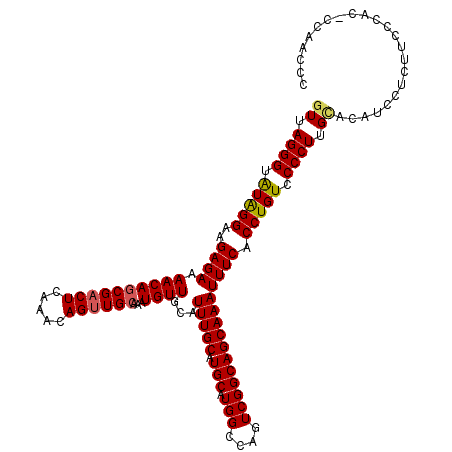
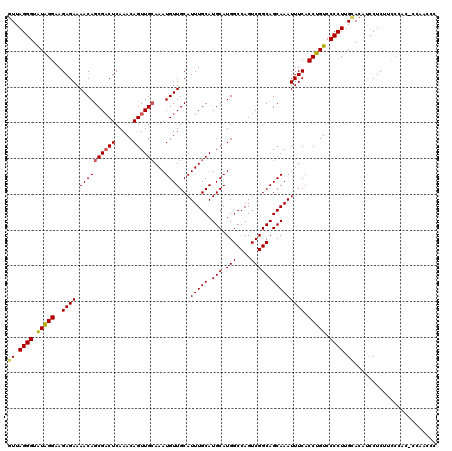
| Location | 17,047,055 – 17,047,173 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.54 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17047055 118 + 22407834 UUCACCUGUCCCCUUGCAC-UCCUCUUCACACCCCAAACCAUCGUCC-UGGCCACAUCCAUCGGCUGGCCAGUUCGGGUGAGCGUUGUCGUUGCCAUUUAGACAAGUGACAUAUGUGCCA .((((.((((.....((..-.........(((((.........(..(-(((((((........).)))))))..))))))((((....))))))......)))).))))........... ( -30.80) >DroSec_CAF1 11154 120 + 1 UUCACCUGUCCCCUUGCACAUCCUCGUUCCACUCCAACCCAUCCCCCAUGGCCCCAUCCAUCGGCUGGCCAGUUCGGGUGAGCGUUGUCGCUGCCAUUUAGACAAGUGACAUAUGCGCCA .(((((((..(..............(((.......)))..........(((((((.......))..))))))..)))))))(((((((((((............))))))).)))).... ( -30.90) >DroSim_CAF1 4810 120 + 1 UUCACCUGUCCCCUUGCACAUCCUCUUUCCACUCCAACCCAUCCCCAAUGGCCCCAUCCAUCGGCUGGCCAGUUCGGGUGAGCGUUGUCGCUGCCAUUUAGACAAGUGACAUAUGCGCCA .((((.((((.....((......................(((((....(((((((.......))..)))))....)))))((((....))))))......)))).))))........... ( -30.30) >DroEre_CAF1 10884 114 + 1 UUCACCUGUCCCCUUGCACCUCCUCUACC-----CCUCCCAUCCCCC-UGGCCCCAUCCAUCGGUUGGCCAGUUCGGGUGAGCGUUGUCGCUGCCAUUUAGACAAGUGACACAUGCGCCA .((((.((((.....((............-----.....(((((..(-(((((((.......))..))))))...)))))((((....))))))......)))).))))........... ( -32.00) >DroYak_CAF1 8789 113 + 1 UUCACCUGUCCCCUCGUACCUCCUCCACC-----CAUGCCAUCCCUC-UGGCCCCAUCCAUCGGUUGGCCAGUUCGGGUG-GCGUUGUCGCUGCCAUUUAGACAAGUGACAUAUGCGCCA ..............((((...........-----.(((((((((..(-(((((((.......))..))))))...)))))-))))(((((((............)))))))..))))... ( -36.20) >consensus UUCACCUGUCCCCUUGCACAUCCUCUUCCCAC_CCAACCCAUCCCCC_UGGCCCCAUCCAUCGGCUGGCCAGUUCGGGUGAGCGUUGUCGCUGCCAUUUAGACAAGUGACAUAUGCGCCA .((((.((((.....(((.....................(((((....(((((((.......))..)))))....))))).(((....))))))......)))).))))........... (-26.46 = -26.54 + 0.08)

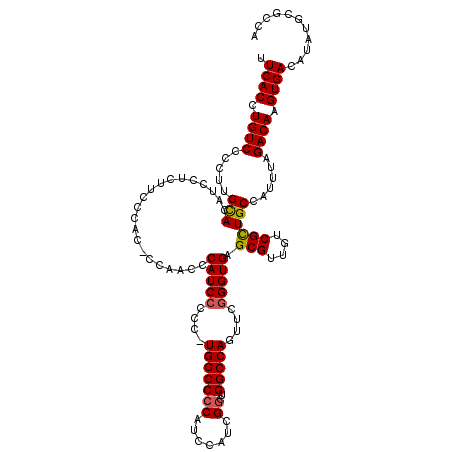
| Location | 17,047,055 – 17,047,173 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -30.44 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17047055 118 - 22407834 UGGCACAUAUGUCACUUGUCUAAAUGGCAACGACAACGCUCACCCGAACUGGCCAGCCGAUGGAUGUGGCCA-GGACGAUGGUUUGGGGUGUGAAGAGGA-GUGCAAGGGGACAGGUGAA ...........((((((((((.....(((.(..(....(.(((((...(((((((.((...))...))))))-)..(((....)))))))).)....)..-))))....)))))))))). ( -43.40) >DroSec_CAF1 11154 120 - 1 UGGCGCAUAUGUCACUUGUCUAAAUGGCAGCGACAACGCUCACCCGAACUGGCCAGCCGAUGGAUGGGGCCAUGGGGGAUGGGUUGGAGUGGAACGAGGAUGUGCAAGGGGACAGGUGAA ...........((((((((((.....(((((..((((.(((.((((...(((((..((...))....))))))))).)).).))))..))............)))....)))))))))). ( -40.80) >DroSim_CAF1 4810 120 - 1 UGGCGCAUAUGUCACUUGUCUAAAUGGCAGCGACAACGCUCACCCGAACUGGCCAGCCGAUGGAUGGGGCCAUUGGGGAUGGGUUGGAGUGGAAAGAGGAUGUGCAAGGGGACAGGUGAA ...........((((((((((.....(((((..((((.(((.(((((..(((((..((...))....)))))))))))).).))))..))............)))....)))))))))). ( -42.40) >DroEre_CAF1 10884 114 - 1 UGGCGCAUGUGUCACUUGUCUAAAUGGCAGCGACAACGCUCACCCGAACUGGCCAACCGAUGGAUGGGGCCA-GGGGGAUGGGAGG-----GGUAGAGGAGGUGCAAGGGGACAGGUGAA ...........((((((((((.....(((.(.((..(.(((((((...((((((..(((.....))))))))-).))).)))).).-----.))......).)))....)))))))))). ( -43.90) >DroYak_CAF1 8789 113 - 1 UGGCGCAUAUGUCACUUGUCUAAAUGGCAGCGACAACGC-CACCCGAACUGGCCAACCGAUGGAUGGGGCCA-GAGGGAUGGCAUG-----GGUGGAGGAGGUACGAGGGGACAGGUGAA ...........((((((((((.....((..(.((...((-(((((...((((((..(((.....))))))))-).))).))))...-----.)).).....))......)))))))))). ( -41.50) >consensus UGGCGCAUAUGUCACUUGUCUAAAUGGCAGCGACAACGCUCACCCGAACUGGCCAGCCGAUGGAUGGGGCCA_GGGGGAUGGGUUGG_GUGGGAAGAGGAGGUGCAAGGGGACAGGUGAA ...........((((((((((.....((((((....))).(((((....(((((..((...))....)))))...))).)).....................)))....)))))))))). (-30.44 = -31.24 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:00 2006