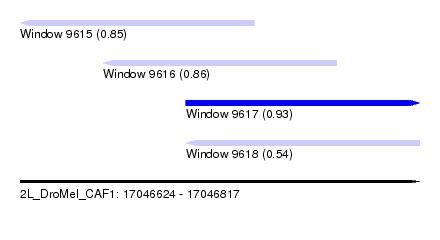
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,046,624 – 17,046,817 |
| Length | 193 |
| Max. P | 0.928025 |

| Location | 17,046,624 – 17,046,737 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17046624 113 - 22407834 ACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGACACCUUGUCUAUGUGUGUUUGCUUAGGCGGGCGAUAAAAA .((...(((-------(((....))))((...((((((.((((((((((((......((((..((.....))...))))....)))).)))))))).))))))...)))).))....... ( -29.70) >DroSec_CAF1 10726 113 - 1 ACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACUACCGGCAGUAAUUUGGACACCUUGUCUAUGUGUGUUUGCUUAGGCGGGCGAUAAAAA .((...(((-------(((....))))((...((((((.((((((((((((......((((.(((.....)))..))))....)))).)))))))).))))))...)))).))....... ( -31.00) >DroSim_CAF1 4382 113 - 1 ACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGACACCUUGUCUAUGUGUGUUUGCUUAGGCGGGCGAUAAAAA .((...(((-------(((....))))((...((((((.((((((((((((......((((..((.....))...))))....)))).)))))))).))))))...)))).))....... ( -29.70) >DroEre_CAF1 10449 120 - 1 ACGACACCAAGCAACGACAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGACACCUUGUCUAUGUGUGUUUGCUUAGGCGGGCGAUAAAAA .((...((.((((..........))))((...((((((.((((((((((((......((((..((.....))...))))....)))).)))))))).))))))...)))).))....... ( -28.80) >DroYak_CAF1 8351 113 - 1 ACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGAAUACACCAACAACCGGCAGUAAUUUGGACACCUUGUCUAUGUGUGUUUGCUUAGGCUGGCGAUAAAAA ....(.(((-------((.....(((((...))))).........((((((.(((((((.......))........(((((.....)))))))))).))))))...))))).)....... ( -28.00) >consensus ACGACACCA_______GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGACACCUUGUCUAUGUGUGUUUGCUUAGGCGGGCGAUAAAAA ....................(((((((.....((((((.((((((((((((......((((..((.....))...))))....)))).)))))))).))))))..)))))).)....... (-27.50 = -27.70 + 0.20)


| Location | 17,046,664 – 17,046,777 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -23.11 |
| Energy contribution | -23.55 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17046664 113 - 22407834 GUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGA (((((((..........((((....))))......)))))))....(((-------(.....(((((((....)).......(((((.....))))).........)))))....)))). ( -27.49) >DroSec_CAF1 10766 113 - 1 GUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACUACCGGCAGUAAUUUGGA (((((((..........((((....))))......)))))))....(((-------(.....(((((((....)).......(((((.....))))).........)))))....)))). ( -27.49) >DroSim_CAF1 4422 113 - 1 GUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCCACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGA (((((...((((.....))))))))).......((((((((((.....(-------(((....))))((....)).......)))))).....(((.((.......))..)))..)))). ( -29.00) >DroEre_CAF1 10489 120 - 1 GUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCAAGCAACGACAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGA (((((((..........((((....))))......)))))))....(((((..((..(.....(((((...)))))......(((((.....)))))..........)..))..))))). ( -26.99) >DroYak_CAF1 8391 113 - 1 GCUGCUUUCGCCAACAAGGUGGCAACACUGACACUAAGCAACGACACCA-------GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGAAUACACCAACAACCGGCAGUAAUUUGGA ((((((...((((.((.((((....))))........((........((-------(((....))))).....)).......))))))..(.......).......))))))........ ( -24.32) >consensus GUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCA_______GCAACCCUGCUGCAACAGCAAAAACAUGUGGCAAGACUACACCAACAACCGGCAGUAAUUUGGA (((((((..........((((....))))......)))))))....(((.......((..((.(((((...)))))......(((((.....))))).........))..))....))). (-23.11 = -23.55 + 0.44)

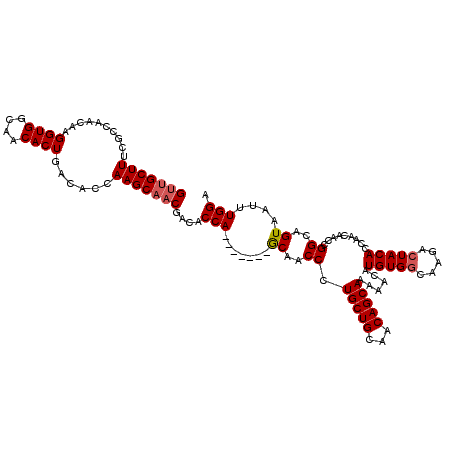
| Location | 17,046,704 – 17,046,817 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -31.18 |
| Energy contribution | -31.06 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17046704 113 + 22407834 UUUUUGCUGUUGCAGCAGGGUUGC-------UGGUGUCGUUGCUUGGUGUCAGUGUUGCCACCUUGUUGGCGAAAGCAACUGCAAAAACAAGAAAAGCUAGAAAAGUCUCUAUAACCGAA (((((((.(((.((((((((((((-------(((..(((.....)))..)))))).....)))))))))((....))))).)))))))..........((((......))))........ ( -33.90) >DroSec_CAF1 10806 113 + 1 UUUUUGCUGUUGCAGCAGGGUUGC-------UGGUGUCGUUGCUUGGUGUCAGUGUUGCCACCUUGUUGGCGAAAGCAACUGCAAAAACAAGAAAAGCUAGAAAAGUCUUCAUAACCGAA (((((((.(((.((((((((((((-------(((..(((.....)))..)))))).....)))))))))((....))))).)))))))...(((..(((.....))).)))......... ( -33.70) >DroSim_CAF1 4462 113 + 1 UUUUUGCUGUUGCAGCAGGGUUGC-------UGGUGUCGUGGCUUGGUGUCAGUGUUGCCACCUUGUUGGCGAAAGCAACUGCAAAAACAAGAAAAGCUAGAAAAGUCUUCAUAACCGAA (((((((.(((.((((((((((((-------(((..(((.....)))..)))))).....)))))))))((....))))).)))))))...(((..(((.....))).)))......... ( -33.70) >DroEre_CAF1 10529 120 + 1 UUUUUGCUGUUGCAGCAGGGUUGUCGUUGCUUGGUGUCGUUGCUUGGUGUCAGUGUUGCCACCUUGUUGGCGAAAGCAACUGCAAAAACAAGAAAAACCAGACUAGCAUCUAUAACCGAA ...((((((...))))))((((((.(.((((.(((.(.(((.((((.((.((((.((((((......)))))).....))))))....))))...))).).))))))).).))))))... ( -38.30) >DroYak_CAF1 8431 113 + 1 UUUUUGCUGUUGCAGCAGGGUUGC-------UGGUGUCGUUGCUUAGUGUCAGUGUUGCCACCUUGUUGGCGAAAGCAGCUGCAAAAACAGGAAAAACAAGAGUAGUUUCUGUAACCGAA .((((((.(((.(((((((((...-------.((((.((((((.....).))))).)))))))))))))((....))))).))))))(((((((..((....))..)))))))....... ( -35.30) >consensus UUUUUGCUGUUGCAGCAGGGUUGC_______UGGUGUCGUUGCUUGGUGUCAGUGUUGCCACCUUGUUGGCGAAAGCAACUGCAAAAACAAGAAAAGCUAGAAAAGUCUCUAUAACCGAA (((((((.(((.(((((((((.((.......(((..(((.....)))..))).....)).)))))))))((....))))).)))))))................................ (-31.18 = -31.06 + -0.12)


| Location | 17,046,704 – 17,046,817 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17046704 113 - 22407834 UUCGGUUAUAGAGACUUUUCUAGCUUUUCUUGUUUUUGCAGUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCA-------GCAACCCUGCUGCAACAGCAAAAA ...((((.(((((....)))))(((.....((((.((((.(((((...((((.....)))))))))..((....)).)))).))))..)-------)))))).(((((...))))).... ( -28.20) >DroSec_CAF1 10806 113 - 1 UUCGGUUAUGAAGACUUUUCUAGCUUUUCUUGUUUUUGCAGUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCA-------GCAACCCUGCUGCAACAGCAAAAA ...(((((.((((...))))))))).......(((((((.(((((((..........((((....))))......))))))).....((-------(((....))))).....))))))) ( -27.69) >DroSim_CAF1 4462 113 - 1 UUCGGUUAUGAAGACUUUUCUAGCUUUUCUUGUUUUUGCAGUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCCACGACACCA-------GCAACCCUGCUGCAACAGCAAAAA ...(((((.((((...))))))))).......(((((((.(((((.............(((((..............)))))......(-------(((....))))))))).))))))) ( -25.64) >DroEre_CAF1 10529 120 - 1 UUCGGUUAUAGAUGCUAGUCUGGUUUUUCUUGUUUUUGCAGUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCAAGCAACGACAACCCUGCUGCAACAGCAAAAA ..........(.(((.(((..((((.((.((((((.((..(((((((..........((((....))))......)))))))..))..)))))).)).))))..)))))).)........ ( -29.49) >DroYak_CAF1 8431 113 - 1 UUCGGUUACAGAAACUACUCUUGUUUUUCCUGUUUUUGCAGCUGCUUUCGCCAACAAGGUGGCAACACUGACACUAAGCAACGACACCA-------GCAACCCUGCUGCAACAGCAAAAA ...((....((((((.......))))))))((((.((((((((((((..........((((....))))......)))))..(....).-------........))))))).)))).... ( -25.09) >consensus UUCGGUUAUAGAGACUUUUCUAGCUUUUCUUGUUUUUGCAGUUGCUUUCGCCAACAAGGUGGCAACACUGACACCAAGCAACGACACCA_______GCAACCCUGCUGCAACAGCAAAAA ...((((.(((((((................)))))))..(((((((..........((((....))))......)))))))................)))).(((((...))))).... (-22.82 = -22.50 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:56 2006