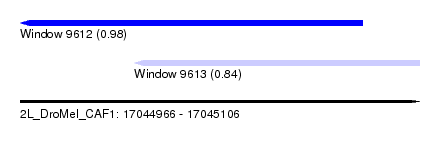
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,044,966 – 17,045,106 |
| Length | 140 |
| Max. P | 0.984012 |

| Location | 17,044,966 – 17,045,086 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -27.38 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17044966 120 - 22407834 UUCUCUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAUUAGCACACACACAAAAUGAAUGCAAAUAUUGGGUUCCAAG ..(((..(((((((((((..(((((((((((((.((((((((......)))).))))...)))))))))))))(((......)))...........)))))).)))))..)))....... ( -31.50) >DroSec_CAF1 9102 120 - 1 UUCUCUUUAUUAUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAUUAGCACACGCACAAAAUGAAUGCAAAUAUUGGGUUCCAAG ..(((..((((.((((((..(((((((((((((.((((((((......)))).))))...)))))))))))))(((......)))...........))))))..))))..)))....... ( -29.50) >DroSim_CAF1 2752 120 - 1 UUCUCUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAUUAGCACACGCACAAAAUGAAUGCAAAUAUUGGGUUCCAAG ..(((..(((((((((((..(((((((((((((.((((((((......)))).))))...)))))))))))))(((......)))...........)))))).)))))..)))....... ( -31.50) >DroEre_CAF1 8899 120 - 1 UUCACUUUGUUUUAUUGGAUUGUGUGGGUGUGUCGCCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAUUAGCACACGCACAAAAUGAAUGCAAAUAUUGGGCUCCAAG ((((.(((((....(((.((.((((((((((((.((..((((......))))..))....)))))))))))))).)))....((....))))))).))))........((((...)))). ( -27.40) >DroYak_CAF1 6648 120 - 1 UUCACUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAUUAGCACACGCACAAAAUGAAUGCAAAUAUUGGGUUCCAAG ....(..(((((((((((..(((((((((((((.((((((((......)))).))))...)))))))))))))(((......)))...........)))))).)))))..)......... ( -29.00) >consensus UUCUCUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAUUAGCACACGCACAAAAUGAAUGCAAAUAUUGGGUUCCAAG ..(((..(((((((((((..(((((((((((((.((((((((......)))).))))...)))))))))))))(((......)))...........)))))).)))))..)))....... (-27.38 = -28.22 + 0.84)

| Location | 17,045,006 – 17,045,106 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17045006 100 - 22407834 U--------------------GAAACUUUCUCUAACCCCUUUCUCUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAU .--------------------....................................(..(((((((((((((.((((((((......)))).))))...)))))))))))))..).... ( -21.10) >DroSec_CAF1 9142 100 - 1 U--------------------GAAACUUUCUCUAACCCCUUUCUCUUUAUUAUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAU .--------------------....................................(..(((((((((((((.((((((((......)))).))))...)))))))))))))..).... ( -21.10) >DroSim_CAF1 2792 100 - 1 U--------------------GAAACUUUCUCUAACCCCUUUCUCUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAU .--------------------....................................(..(((((((((((((.((((((((......)))).))))...)))))))))))))..).... ( -21.10) >DroEre_CAF1 8939 109 - 1 UCGAGAAGUA-----------GAAACUUUCUCCAACCCCUUUCACUUUGUUUUAUUGGAUUGUGUGGGUGUGUCGCCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAU ..(((((((.-----------...)).)))))......................(((.((.((((((((((((.((..((((......))))..))....)))))))))))))).))).. ( -24.10) >DroYak_CAF1 6688 120 - 1 UCGAAAAGUCGGGAGUGAGGCGAAACUUUCUCUAACCCCUUUCACUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAU (((((.((...(((((((((.(...............).)))))))))...)).))))).(((((((((((((.((((((((......)))).))))...)))))))))))))....... ( -31.56) >consensus U____________________GAAACUUUCUCUAACCCCUUUCUCUUUAUUUUAUUCGAUUGUGUGGGUGUGUCGGCAGAGUCAACUAAUUUAUGUUUUAACACACUCGCACAUGCAAAU .........................................................(..(((((((((((((.((((((((......)))).))))...)))))))))))))..).... (-20.24 = -20.44 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:52 2006