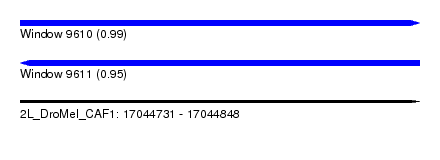
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,044,731 – 17,044,848 |
| Length | 117 |
| Max. P | 0.992292 |

| Location | 17,044,731 – 17,044,848 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17044731 117 + 22407834 GAAUACCGCAAAUUGACAAAUAUUUUCGUUUGGCCAACAAGAAGAAGACCAAAAGCA---CCCAGCACUCUAAUUCAAUCUGGCUGGUGUAUUUUAAUUGAUUUGCGCUAAAAUUUAUUG ...((.(((((((..(......(((((.((((.....))))..)))))......(((---((.(((.(.............))))))))).......)..))))))).)).......... ( -22.72) >DroSec_CAF1 8865 117 + 1 GAAUACCGCAAAUUGACAAAUAUUUUCGUUUGGCCAACAAGAAGAAGGCCAAAAGCA---CCCAGCACUCUAAUUCAAUCUGGCUGGUGUAUUUUAAUUGAUUUGCGCUAAAAUUUAUUG ...((.(((((((..(............(((((((..(.....)..))))))).(((---((.(((.(.............))))))))).......)..))))))).)).......... ( -29.12) >DroSim_CAF1 2512 120 + 1 GAAUACCGCAAAUUGACAAAUAUUUUCGUUUGGCCAACAAGAAGAAGGCCAAAAGCACCACCCAGCACUCUAAUUCAAUCUGGCUGGUGUAUUUUAAUUGAUUUGCGCUAAAAUUUAUUG ...((.(((((((..(............(((((((..(.....)..))))))).((((((.((((..............)))).)))))).......)..))))))).)).......... ( -32.74) >DroEre_CAF1 8669 116 + 1 GCAUACCGCAAAUGGACAAAUAUUUUCGUUUGGCCAACAAGAAGAAGGCCAAAAGCA---CCCAGCACUCUAAUUCAAUCUGGCUGGUGUAUUUUAAUUGAUUUGCGCUAA-AUUUAUUG ...((.(((((((.((............(((((((..(.....)..))))))).(((---((.(((.(.............))))))))).......)).))))))).)).-........ ( -27.92) >DroYak_CAF1 6415 116 + 1 GAAUACCGCAAAUCGACAAAUAUUUUUGUUUGGCCAACAAAAAGAAGACCAAAAGCA---CCCAGCACUCUAAUUCAAUCUGGCUGGGGUAUUUUAAUUGAUUUGCGCUAA-AUUUAUUG ...((.((((((((((......((((((((.....)))))))).......((((...---((((((.(.............)))))))...))))..)))))))))).)).-........ ( -27.92) >consensus GAAUACCGCAAAUUGACAAAUAUUUUCGUUUGGCCAACAAGAAGAAGGCCAAAAGCA___CCCAGCACUCUAAUUCAAUCUGGCUGGUGUAUUUUAAUUGAUUUGCGCUAAAAUUUAUUG ...((.((((((((((.((((((.....(((((((..(.....)..)))))))........(((((.(.............)))))).))))))...)))))))))).)).......... (-24.30 = -24.78 + 0.48)


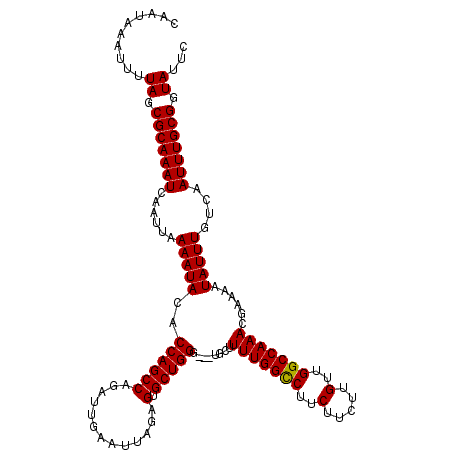
| Location | 17,044,731 – 17,044,848 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17044731 117 - 22407834 CAAUAAAUUUUAGCGCAAAUCAAUUAAAAUACACCAGCCAGAUUGAAUUAGAGUGCUGGG---UGCUUUUGGUCUUCUUCUUGUUGGCCAAACGAAAAUAUUUGUCAAUUUGCGGUAUUC ..........((.((((((((((.((...(((.((((((.............).))))))---))..(((((((..(.....)..)))))))......)).)))...))))))).))... ( -24.12) >DroSec_CAF1 8865 117 - 1 CAAUAAAUUUUAGCGCAAAUCAAUUAAAAUACACCAGCCAGAUUGAAUUAGAGUGCUGGG---UGCUUUUGGCCUUCUUCUUGUUGGCCAAACGAAAAUAUUUGUCAAUUUGCGGUAUUC ..........((.((((((((((.((...(((.((((((.............).))))))---))..(((((((..(.....)..)))))))......)).)))...))))))).))... ( -26.82) >DroSim_CAF1 2512 120 - 1 CAAUAAAUUUUAGCGCAAAUCAAUUAAAAUACACCAGCCAGAUUGAAUUAGAGUGCUGGGUGGUGCUUUUGGCCUUCUUCUUGUUGGCCAAACGAAAAUAUUUGUCAAUUUGCGGUAUUC ..........((.(((((((......((((((((((.((((.((......))...)))).)))))..(((((((..(.....)..)))))))......)))))....))))))).))... ( -34.00) >DroEre_CAF1 8669 116 - 1 CAAUAAAU-UUAGCGCAAAUCAAUUAAAAUACACCAGCCAGAUUGAAUUAGAGUGCUGGG---UGCUUUUGGCCUUCUUCUUGUUGGCCAAACGAAAAUAUUUGUCCAUUUGCGGUAUGC ........-.((.((((((((((.((...(((.((((((.............).))))))---))..(((((((..(.....)..)))))))......)).)))...))))))).))... ( -26.82) >DroYak_CAF1 6415 116 - 1 CAAUAAAU-UUAGCGCAAAUCAAUUAAAAUACCCCAGCCAGAUUGAAUUAGAGUGCUGGG---UGCUUUUGGUCUUCUUUUUGUUGGCCAAACAAAAAUAUUUGUCGAUUUGCGGUAUUC ........-.((.((((((((...........(((((((.............).))))))---....(((((((..(.....)..)))))))..............)))))))).))... ( -29.72) >consensus CAAUAAAUUUUAGCGCAAAUCAAUUAAAAUACACCAGCCAGAUUGAAUUAGAGUGCUGGG___UGCUUUUGGCCUUCUUCUUGUUGGCCAAACGAAAAUAUUUGUCAAUUUGCGGUAUUC ..........((.(((((((......(((((..((((((.............).)))))........(((((((..(.....)..)))))))......)))))....))))))).))... (-25.44 = -25.20 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:50 2006