
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,031,282 – 17,031,372 |
| Length | 90 |
| Max. P | 0.621652 |

| Location | 17,031,282 – 17,031,372 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.44 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
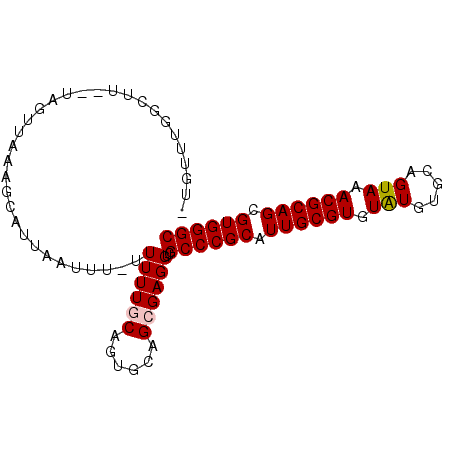
>2L_DroMel_CAF1 17031282 90 + 22407834 -UGUUUGGCUUGAUAGUUAAAGCUUCAAUUU-UUUUUACAGUGCAGCGAGGUCGCCCGCAUUGCGUGUAUGUGCAGUAAACGCAGCGUGGGC -..(((((((....)))))))(((.((....-.........)).)))......((((((.((((((.(((.....))).)))))).)))))) ( -25.52) >DroSec_CAF1 138672 81 + 1 -AGUGUGUAUU----------GUAUAAAUUUUUUUUUGCAGUGCAGCGAGGUCGCCCGCAUUGCGUGUAUGUGCAGUAAACGCAGCGUGGGC -.((.((((((----------(((.(((....))).)))))))))))......((((((.((((((.(((.....))).)))))).)))))) ( -30.20) >DroSim_CAF1 141836 89 + 1 -UUUUUGGCUUGAUAGUUAAAGCAUGAAUUU--UUUUACAGUGCAGCGAGGUCGCCCGCAUUGCGUGUAUGUGCAGUAAACGCAGCGUGGGC -..(((((((....)))))))((((.((...--..))...)))).........((((((.((((((.(((.....))).)))))).)))))) ( -26.90) >DroEre_CAF1 140151 88 + 1 AUGUUUGCUUG--UAGUUAUUACCUUG-CAU-UUUUUGCAGUGCAGCGAGGUCGCCCGCAUUGCGUGUGUGUGCAGUAAACGCAGCGUGGGC ..(((..((((--(.(((..(((..((-(((-....((((((((.(((....)))..)))))))).....))))))))))))))).)..))) ( -28.60) >DroYak_CAF1 145247 89 + 1 AUGUUUGGCUG--CAUGUUUUGCCUUGACAU-UUUUUGCAGUGCAGCGAGGUCGCCCGCAUUGCGUGUGUGUGCAGUAAACGCAGCGUGGGC ..((..(((((--(.....((((....((((-....((((((((.(((....)))..))))))))...)))))))).....))))).)..)) ( -30.90) >consensus _UGUUUGGCUU__UAGUUAAAGCAUUAAUUU_UUUUUGCAGUGCAGCGAGGUCGCCCGCAUUGCGUGUAUGUGCAGUAAACGCAGCGUGGGC .................................((((((......))))))..((((((.((((((.(((.....))).)))))).)))))) (-20.28 = -20.44 + 0.16)

| Location | 17,031,282 – 17,031,372 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -16.50 |
| Energy contribution | -16.26 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17031282 90 - 22407834 GCCCACGCUGCGUUUACUGCACAUACACGCAAUGCGGGCGACCUCGCUGCACUGUAAAAA-AAAUUGAAGCUUUAACUAUCAAGCCAAACA- ((((.((.(((((.((.......)).))))).)).)))).....................-........((((........))))......- ( -17.60) >DroSec_CAF1 138672 81 - 1 GCCCACGCUGCGUUUACUGCACAUACACGCAAUGCGGGCGACCUCGCUGCACUGCAAAAAAAAAUUUAUAC----------AAUACACACU- ((((.((.(((((.((.......)).))))).)).))))......((......))................----------..........- ( -17.70) >DroSim_CAF1 141836 89 - 1 GCCCACGCUGCGUUUACUGCACAUACACGCAAUGCGGGCGACCUCGCUGCACUGUAAAA--AAAUUCAUGCUUUAACUAUCAAGCCAAAAA- ((((.((.(((((.((.......)).))))).)).))))....................--........((((........))))......- ( -17.40) >DroEre_CAF1 140151 88 - 1 GCCCACGCUGCGUUUACUGCACACACACGCAAUGCGGGCGACCUCGCUGCACUGCAAAAA-AUG-CAAGGUAAUAACUA--CAAGCAAACAU ((((.((.(((((.............))))).)).))))......(((....((((....-.))-))..(((.....))--).)))...... ( -21.42) >DroYak_CAF1 145247 89 - 1 GCCCACGCUGCGUUUACUGCACACACACGCAAUGCGGGCGACCUCGCUGCACUGCAAAAA-AUGUCAAGGCAAAACAUG--CAGCCAAACAU ......(((((((....(((..(((...(((.(((((.((....))))))).))).....-.)))....)))....)))--))))....... ( -25.10) >consensus GCCCACGCUGCGUUUACUGCACAUACACGCAAUGCGGGCGACCUCGCUGCACUGCAAAAA_AAAUUAAGGCAAUAACUA__AAGCCAAACA_ ((((.((.(((((.............))))).)).))))......((......))..................................... (-16.50 = -16.26 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:44 2006