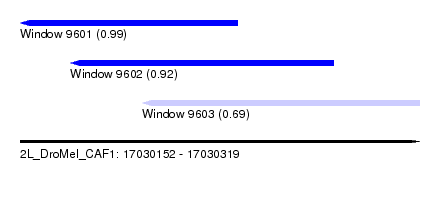
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,030,152 – 17,030,319 |
| Length | 167 |
| Max. P | 0.989780 |

| Location | 17,030,152 – 17,030,243 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.06 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -30.67 |
| Energy contribution | -30.55 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17030152 91 - 22407834 UGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAAC------GC------------GAGCGCGCUUCAGACAAAUUA .(((((((((((.(((((....))))).)))).))))))).......(((.(..((((((..((....------))------------..))))))..).)))...... ( -35.30) >DroSec_CAF1 137468 97 - 1 UGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCAAACGC------------GAACGCGCUUCAAACAAAUUA .(((((((((((.(((((....))))).)))).)))))))....(((((..(((.(((((..((....))..))))------------))))..))))).......... ( -36.40) >DroSim_CAF1 140702 97 - 1 UGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCAAACGC------------GAACGCGCUUCAGACAAAUUA .(((((((((((.(((((....))))).)))).))))))).......(((.(..((((((..(((........)))------------..))))))..).)))...... ( -36.50) >DroEre_CAF1 139021 97 - 1 UGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGACGUUAGUUGCGUGUGAGCGAACGCAAACGC------------GAACGCGCUGCAGACAAAUUA .(((((((((((.(((((....))))).)))).))))))).......(((.((.((((((..(((........)))------------..)))))).)).)))...... ( -38.80) >DroYak_CAF1 144024 109 - 1 UGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCGAACGCAAACGCGAACGGGAACGCGCUUCAGACAAAUUA .(((((((((((.(((((....))))).)))).))))))).......(((.(..((((((...((..((((........))))..))...))))))..).)))...... ( -38.20) >consensus UGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCAAACGC____________GAACGCGCUUCAGACAAAUUA .(((((((((((.(((((....))))).)))).))))))).......(((.(..((((((..(((........)))..............))))))..).)))...... (-30.67 = -30.55 + -0.12)

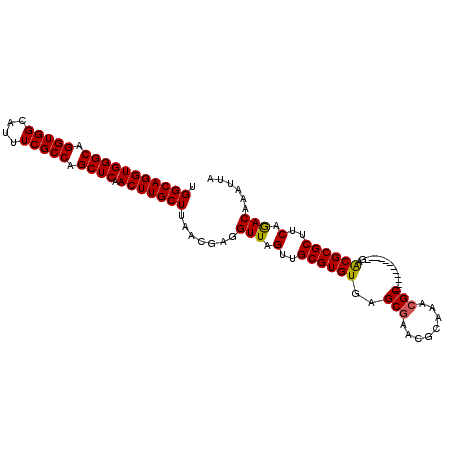
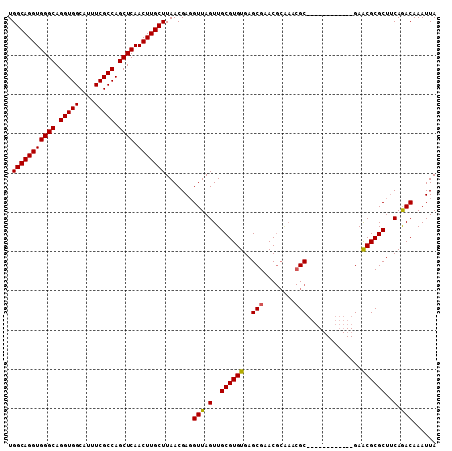
| Location | 17,030,173 – 17,030,283 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -43.34 |
| Consensus MFE | -39.88 |
| Energy contribution | -40.76 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17030173 110 - 22407834 CAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAAC------GC--- ...((.(((.((((((((((((((....)))))....((..(((((((((((.(((((....))))).)))).)))))))..)).......)))))))))..))).))------..--- ( -39.60) >DroSec_CAF1 137489 116 - 1 CAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCAAACGC--- .....((((....((((...((..((.((((.(((...((((((((((((((.(((((....))))).)))).)))))))..)))...))).)))).))..))....)))).))))--- ( -43.10) >DroSim_CAF1 140723 116 - 1 CAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCAAACGC--- .....((((....((((...((..((.((((.(((...((((((((((((((.(((((....))))).)))).)))))))..)))...))).)))).))..))....)))).))))--- ( -43.10) >DroEre_CAF1 139042 116 - 1 CAGGCGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAACUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGACGUUAGUUGCGUGUGAGCGAACGCAAACGC--- ...((((((.(((((((((........((((......))))(((((((((((.(((((....))))).)))).)))))))...........)))))))))..))..))))......--- ( -43.20) >DroYak_CAF1 144054 119 - 1 CAGGUGUGUGGCGUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCGAACGCAAA ....(((((.((((.((...((..((.((((.(((...((((((((((((((.(((((....))))).)))).)))))))..)))...))).)))).))..)))).))))..))))).. ( -47.70) >consensus CAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCUUAACGAGGUUAGUUGCGUGUGAGCGAACGCAAACGC___ .....((((....((((...((..((.((((.(((...((((((((((((((.(((((....))))).)))).)))))))..)))...))).)))).))..))....)))).))))... (-39.88 = -40.76 + 0.88)



| Location | 17,030,203 – 17,030,319 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -44.26 |
| Consensus MFE | -43.42 |
| Energy contribution | -43.02 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17030203 116 - 22407834 GCAACUUGCAAGUGGCAGCUCAGCUAAGAGUCAGGACAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCU ((..((((((((((.(.((((......))))........(((((...)))))).))))))))))...))........(((((((((((.(((((....))))).)))).))))))) ( -40.70) >DroSec_CAF1 137525 116 - 1 GCAACUUGCAAGUGGCAGCUCAGCUAGGAGUCAGGACAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCU ((..((((((((((.(.((...((((.(..((......))..).))))..))).))))))))))...))........(((((((((((.(((((....))))).)))).))))))) ( -42.60) >DroSim_CAF1 140759 116 - 1 GCAACUUGCAGGUGGCAGCUCAGCUAGGAGUCAGGACAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCU ((..((((((((((.(.((...((((.(..((......))..).))))..))).))))))))))...))........(((((((((((.(((((....))))).)))).))))))) ( -42.20) >DroEre_CAF1 139078 116 - 1 GCAACUUGCAAGUGGCAGCUCUGCCAGGAGUCAGGACAGGCGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAACUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCU ....((((((((((.(.((..(((((.(.(((......))).).))))).))).))))))))))(((......))).(((((((((((.(((((....))))).)))).))))))) ( -49.80) >DroYak_CAF1 144093 116 - 1 GCAACUUGCAAGUGGCAGCUCAGCCAGGAGUCAGGACAGGUGUGUGGCGUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCU ((..((((((((((.(.((...((((.(..((......))..).))))..))).))))))))))...))........(((((((((((.(((((....))))).)))).))))))) ( -46.00) >consensus GCAACUUGCAAGUGGCAGCUCAGCUAGGAGUCAGGACAGGUGUGUGGCAUGCGGCAUUUGCAGGCAGGUGAAAGUGUGGCAGGUGGGCAGGUGGCAUUUCGCCAGCUCAACUUGCU ((..((((((((((.(.((...((((.(..((......))..).))))..))).))))))))))...))........(((((((((((.(((((....))))).)))).))))))) (-43.42 = -43.02 + -0.40)


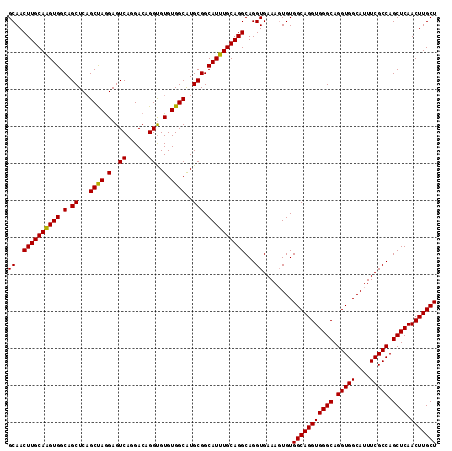
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:42 2006