| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,023,938 – 17,024,066 |
| Length | 128 |
| Max. P | 0.899784 |

| Location | 17,023,938 – 17,024,046 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -26.35 |
| Energy contribution | -25.55 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17023938 108 + 22407834 ACUGAAGGCCAAUCUGGUUGGACUUUUAUUGAGGAAAUUAUACGAGUCUAGGUAGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUGUGGCAGCACCUGUUUUGUCC ......(((((.((((.((((((((.(((..........))).)))))))).)))).((((........))))..)))))(((((.(((....))))))))....... ( -31.20) >DroSec_CAF1 131597 108 + 1 ACUGAAGGCCAAUCUGGUUGGACUUUUAUUGAGGAAAUUAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUGUGGCAGCACCUGUUUUGUCC ......(((((.((((.((((((((.(((..........))).)))))))).)))).((((........))))..)))))(((((.(((....))))))))....... ( -29.90) >DroSim_CAF1 134630 108 + 1 ACUGAAGGCCAAUCUGGUUGGACUUUUAUUGAGGAAAUUAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUGUGGCAGCACCUGUUUUGUCC ......(((((.((((.((((((((.(((..........))).)))))))).)))).((((........))))..)))))(((((.(((....))))))))....... ( -29.90) >DroEre_CAF1 133137 108 + 1 ACUGAAGGCCAAUCUGGUUGGACUUUUAUUGAGGAAAUUAUACGAGUCGAGGUGGAAAACAAAUGUUAAUGUUGCUGGCCACAGGAUAUGGCAGCACCUGCUUUGUCC ......(((((.((((.((.(((((.(((..........))).))))).)).)))).((((........))))..)))))...(((((.(((((...))))).))))) ( -30.60) >DroYak_CAF1 137991 108 + 1 ACUGAAGGCCAAUCUGGUUGGACUUUUAUUGAGGAAAUUAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUGCUGGCCACAGGAUAUGGCAGCACCUGUUUUGUUC ......(((((.((((.((((((((.(((..........))).)))))))).)))).((((........))))..)))))((((((((.((.....)))))))))).. ( -28.20) >DroAna_CAF1 123332 105 + 1 ---GAAAGCUAAUCUGGUUGGACUUUUAUUGAGGAAAUUGUACAAGUGUAGGUGGAAAGCGAAUGUUAAUGUUUCUGGCCACAGGAUAUGGCGGCACCUGUUUCCUUU ---((((((((((...))))).)))))...(((((((((((....((((..((((..((.((((......))))))..))))...))))....)))...)))))))). ( -24.80) >consensus ACUGAAGGCCAAUCUGGUUGGACUUUUAUUGAGGAAAUUAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUAUGGCAGCACCUGUUUUGUCC ......(((((.((((.((((((((.(((..........))).)))))))).)))).((((........))))..)))))...(((((.(((((...))))).))))) (-26.35 = -25.55 + -0.80)

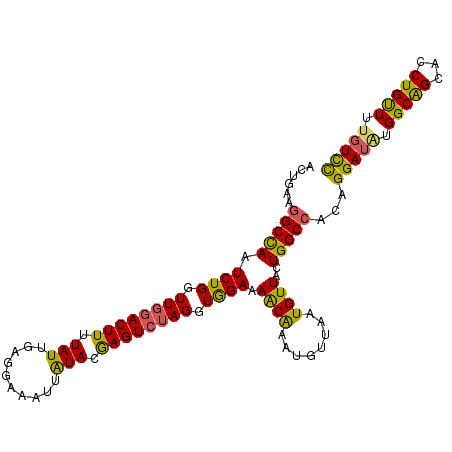
| Location | 17,023,976 – 17,024,066 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.03 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17023976 90 + 22407834 UAUACGAGUCUAGGUAGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUGUGGCAGCACCUGUUUUGUCCACCUUCAC---------CUCCGCUUUUCC----------- .....(((...(((((.((((((..((((..........(((((.....)))))))))..)))))).)..))))....---------))).........----------- ( -18.70) >DroSec_CAF1 131635 90 + 1 UAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUGUGGCAGCACCUGUUUUGUCCACCUCCAC---------CUGUGAUUUUCC----------- ..((((.((..((((((((((((..((((..........(((((.....)))))))))..)))))..)))))))..))---------.)))).......----------- ( -26.10) >DroSim_CAF1 134668 90 + 1 UAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUGUGGCAGCACCUGUUUUGUCCACCUCCAC---------CUCUGAUUUUCC----------- .....(((...((((((((((((..((((..........(((((.....)))))))))..)))))..)))))))....---------))).........----------- ( -24.90) >DroEre_CAF1 133175 99 + 1 UAUACGAGUCGAGGUGGAAAACAAAUGUUAAUGUUGCUGGCCACAGGAUAUGGCAGCACCUGCUUUGUCCACCUCCACUUUCUCCACUUCCUCCUUUGC----------- .....((((.((((((((...((((.((...((((((((.((...))...))))))))...)))))))))))))).))))...................----------- ( -29.40) >DroYak_CAF1 138029 84 + 1 UAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUGCUGGCCACAGGAUAUGGCAGCACCUGUUUUGUUCACCUCCAG---------------CUUUCC----------- .....(((.((.((.((..((((((......((((((((.((...))...)))))))).....))))))..)).))))---------------)))...----------- ( -20.60) >DroAna_CAF1 123367 101 + 1 UGUACAAGUGUAGGUGGAAAGCGAAUGUUAAUGUUUCUGGCCACAGGAUAUGGCGGCACCUGUUUCCUUUUCCUGCUC---------CUGCUCCUGCUCCUGCCCCAUCC .(((..(((((((((((..((.((((......))))))..)))(((((...((..((....))..))...)))))..)---------))))....)))..)))....... ( -23.80) >consensus UAUACGAGUCUAGGUGGAAAACAAAUGUUAAUGUUACUGGCCACAGGAUAUGGCAGCACCUGUUUUGUCCACCUCCAC_________CUCCGCCUUUCC___________ ...........((((((((((((..((((.(((((.(((....))))))))...))))..)))))..))))))).................................... (-17.50 = -17.03 + -0.47)
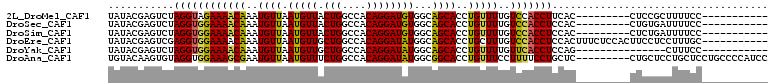

| Location | 17,023,976 – 17,024,066 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -19.15 |
| Energy contribution | -19.85 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17023976 90 - 22407834 -----------GGAAAAGCGGAG---------GUGAAGGUGGACAAAACAGGUGCUGCCACAUCCUGUGGCCAGUAACAUUAACAUUUGUUUUCUACCUAGACUCGUAUA -----------......(((..(---------((..(((((((..((((((((((((((((.....)))).))).........))))))))))))))))..))))))... ( -27.10) >DroSec_CAF1 131635 90 - 1 -----------GGAAAAUCACAG---------GUGGAGGUGGACAAAACAGGUGCUGCCACAUCCUGUGGCCAGUAACAUUAACAUUUGUUUUCCACCUAGACUCGUAUA -----------........((..---------((..(((((((..((((((((((((((((.....)))).))).........))))))))))))))))..))..))... ( -30.00) >DroSim_CAF1 134668 90 - 1 -----------GGAAAAUCAGAG---------GUGGAGGUGGACAAAACAGGUGCUGCCACAUCCUGUGGCCAGUAACAUUAACAUUUGUUUUCCACCUAGACUCGUAUA -----------.........(((---------....(((((((..((((((((((((((((.....)))).))).........))))))))))))))))...)))..... ( -30.30) >DroEre_CAF1 133175 99 - 1 -----------GCAAAGGAGGAAGUGGAGAAAGUGGAGGUGGACAAAGCAGGUGCUGCCAUAUCCUGUGGCCAGCAACAUUAACAUUUGUUUUCCACCUCGACUCGUAUA -----------....................(((.((((((((..((((((((((((((((.....)))).))))).........))))))))))))))).)))...... ( -31.60) >DroYak_CAF1 138029 84 - 1 -----------GGAAAG---------------CUGGAGGUGAACAAAACAGGUGCUGCCAUAUCCUGUGGCCAGCAACAUUAACAUUUGUUUUCCACCUAGACUCGUAUA -----------.((...---------------((((.((.(((((((.....(((((((((.....)))).))))).........))))))).)).)).))..))..... ( -21.84) >DroAna_CAF1 123367 101 - 1 GGAUGGGGCAGGAGCAGGAGCAG---------GAGCAGGAAAAGGAAACAGGUGCCGCCAUAUCCUGUGGCCAGAAACAUUAACAUUCGCUUUCCACCUACACUUGUACA (..((((...(((....((((.(---------(.((((((...(....).((.....))...))))))..)).(((.........)))))))))).))))..)....... ( -26.40) >consensus ___________GGAAAAGCGGAG_________GUGGAGGUGGACAAAACAGGUGCUGCCACAUCCUGUGGCCAGUAACAUUAACAUUUGUUUUCCACCUAGACUCGUAUA ....................................(((((((..((((((((((((((((.....)))).))).........))))))))))))))))........... (-19.15 = -19.85 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:35 2006