
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,009,654 – 17,009,760 |
| Length | 106 |
| Max. P | 0.769504 |

| Location | 17,009,654 – 17,009,760 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.35 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17009654 106 + 22407834 --CCAUUC--CCGCUCGUGUAUCGUUUUGCAAGCUUGAGCGAAAAUGCAAAGAGCUGAAAAAUGCAAUAAGUAAAACGUAAUAACU---GCUCAGGAGCUGUGAACGGUUGGC --((.(((--(.((((.((....(((((((.(((((..((......))...)))))(.......).....)))))))(((.....)---)).)).)))).).))).))..... ( -23.60) >DroPse_CAF1 132155 98 + 1 CAGCAGUC--CUGGUCCUGUAUCGUUUUGCAAGCUCCAGUGCAAAUGCAAAGAGCGCAAAAAUGCAAUAAGUAAAAUGUAAUAACU---GCUCAGGACUCAGG---------- .......(--(((((((((...((((((((..((((...(((....)))..))))(((....))).....))))))))........---...)))))).))))---------- ( -32.84) >DroEre_CAF1 119411 106 + 1 --CCAUUC--CCGCUCCUGUAUCGUUUUGCAAGCUCGAGCGAAAAUGCAAAGAGCUGAAAAAUGCAAUAAGUAAAACGUAAUAACU---GCUCAGGAGCUGUGAACGGUUGGC --((.(((--(.(((((((....(((((((.(((((..((......))...)))))(.......).....)))))))(((.....)---)).))))))).).))).))..... ( -29.80) >DroYak_CAF1 119755 106 + 1 --CCUUGC--CAGCUCCUGUAUCGCUUUGCAAGCUCGAGCGAAAAUGCAAAGAGCUGAAAAAUGCAAUAAGUAAAACGUAAUAACU---GCUCAGGAGCUGUGAACGGUUGGC --((..((--(((((((((....(((((((((((((..((......))...)))))......))))..)))).....(((.....)---)).))))))))(....)))).)). ( -30.70) >DroAna_CAF1 110192 111 + 1 --CCAUUGCACCUCUCCCAUAUCGUUUUGCAAGCUCUAGCGAAAAUGCAAAGAGCAGAAAAAUGCAAUAAGUAAAACGUAAUAACUGGAGCUCAGGAGCCGAGAACGCUUGCC --.....((.(((((((.....((((((((..(((((.((......))..))))).(.......).....))))))))........))))...))).))((((....)))).. ( -24.12) >DroPer_CAF1 141530 98 + 1 CAGCAGUC--CUGGUCCUGUAUCGUUUUGCAAGCUCCAGUGCAAAUGCAAAGAGCGCAAAAAUGCAAUAAGUAAAAUGUAAUAACU---GCUCAGGACUCAGG---------- .......(--(((((((((...((((((((..((((...(((....)))..))))(((....))).....))))))))........---...)))))).))))---------- ( -32.84) >consensus __CCAUUC__CCGCUCCUGUAUCGUUUUGCAAGCUCGAGCGAAAAUGCAAAGAGCUGAAAAAUGCAAUAAGUAAAACGUAAUAACU___GCUCAGGAGCCGUGAACGGUUGGC ............(((((((...((((((((..((((..((......))...)))).......(......)))))))))..............))))))).............. (-18.70 = -18.35 + -0.36)

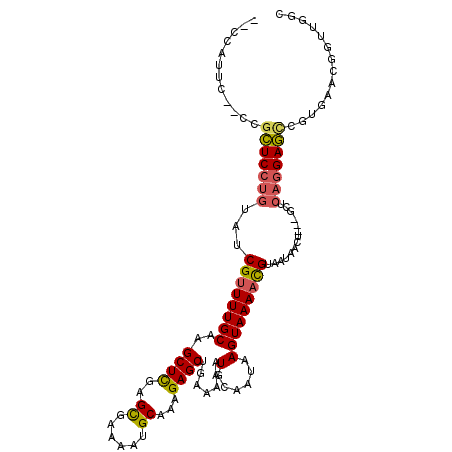
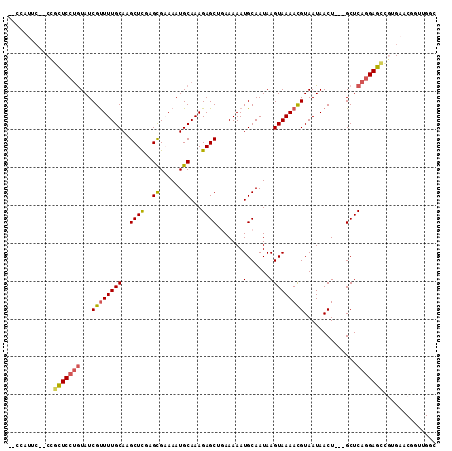
| Location | 17,009,654 – 17,009,760 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.42 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17009654 106 - 22407834 GCCAACCGUUCACAGCUCCUGAGC---AGUUAUUACGUUUUACUUAUUGCAUUUUUCAGCUCUUUGCAUUUUCGCUCAAGCUUGCAAAACGAUACACGAGCGG--GAAUGG-- .....((((((...((((.((.((---(((...............)))))............((((((.....((....)).))))))......)).))))..--))))))-- ( -24.26) >DroPse_CAF1 132155 98 - 1 ----------CCUGAGUCCUGAGC---AGUUAUUACAUUUUACUUAUUGCAUUUUUGCGCUCUUUGCAUUUGCACUGGAGCUUGCAAAACGAUACAGGACCAG--GACUGCUG ----------((((.((((((.((---(((...............)))))..(((((((((((.(((....)))..)))))..)))))).....)))))))))--)....... ( -34.96) >DroEre_CAF1 119411 106 - 1 GCCAACCGUUCACAGCUCCUGAGC---AGUUAUUACGUUUUACUUAUUGCAUUUUUCAGCUCUUUGCAUUUUCGCUCGAGCUUGCAAAACGAUACAGGAGCGG--GAAUGG-- .....((((((...(((((((.((---(((...............))))).((((.((((((...((......))..)))).)).)))).....)))))))..--))))))-- ( -33.56) >DroYak_CAF1 119755 106 - 1 GCCAACCGUUCACAGCUCCUGAGC---AGUUAUUACGUUUUACUUAUUGCAUUUUUCAGCUCUUUGCAUUUUCGCUCGAGCUUGCAAAGCGAUACAGGAGCUG--GCAAGG-- .....((.....(((((((((.((---(((...............))))).........(.(((((((..(((....)))..))))))).)...)))))))))--....))-- ( -33.76) >DroAna_CAF1 110192 111 - 1 GGCAAGCGUUCUCGGCUCCUGAGCUCCAGUUAUUACGUUUUACUUAUUGCAUUUUUCUGCUCUUUGCAUUUUCGCUAGAGCUUGCAAAACGAUAUGGGAGAGGUGCAAUGG-- (..((((((.((.((((....))).).)).....))))))..).(((((((((((((((((((..((......)).)))))(((.....)))....)))))))))))))).-- ( -30.10) >DroPer_CAF1 141530 98 - 1 ----------CCUGAGUCCUGAGC---AGUUAUUACAUUUUACUUAUUGCAUUUUUGCGCUCUUUGCAUUUGCACUGGAGCUUGCAAAACGAUACAGGACCAG--GACUGCUG ----------((((.((((((.((---(((...............)))))..(((((((((((.(((....)))..)))))..)))))).....)))))))))--)....... ( -34.96) >consensus GCCAACCGUUCACAGCUCCUGAGC___AGUUAUUACGUUUUACUUAUUGCAUUUUUCAGCUCUUUGCAUUUUCGCUCGAGCUUGCAAAACGAUACAGGAGCGG__GAAUGG__ ...............((((((.((...(((...........)))....))............((((((..(((....)))..))))))......))))))............. (-15.47 = -15.42 + -0.05)

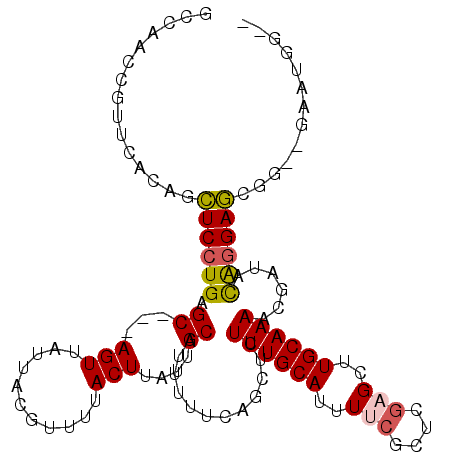
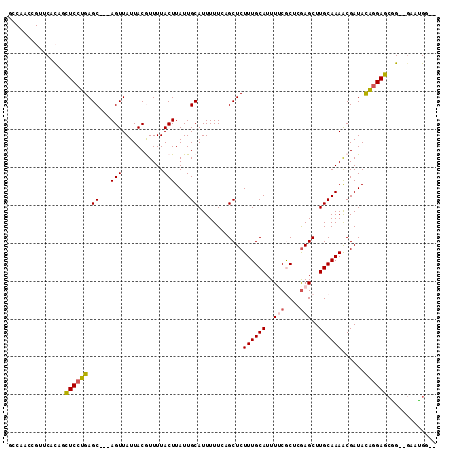
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:29 2006