| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
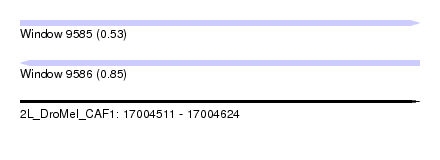
| Location | 17,004,511 – 17,004,624 |
| Length | 113 |
| Max. P | 0.850331 |

| Location | 17,004,511 – 17,004,624 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -13.94 |
| Energy contribution | -15.13 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.37 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
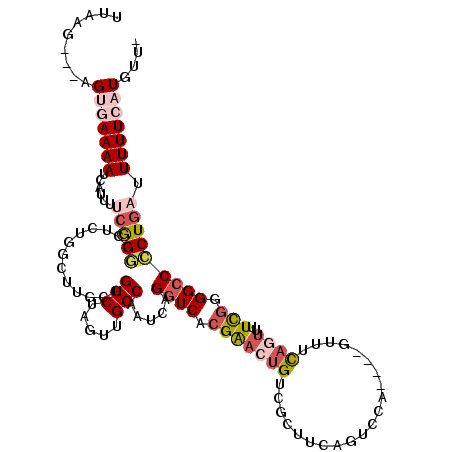
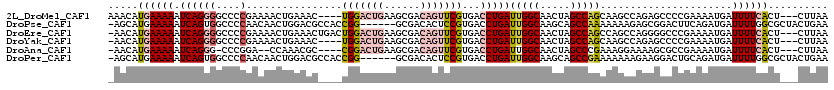
>2L_DroMel_CAF1 17004511 113 + 22407834 UUAAG---AGUGAAAAUCAUUUUCGGGGCUCUGGCUUGCUGGCUAGUUGCCAAUCAGGUCACGAACUGUCGCUUCAGUCCA----GUUUCAGUUUUCGGGGCCCCUGAUUUUUCAUGUUU .....---.((((((((((.....((((((((((((((.((((.....))))..))))))..((((((..(((.......)----))..))))))..))))))))))).))))))).... ( -40.20) >DroPse_CAF1 124906 113 + 1 UUCAGUAGCGCCAAAAUCAUCUGAAGUCCGCUCUUUUUUUGGCUGCUUGCCAAUCAGGUCACGGAGUGUCGC------CCGGUGGCGUCCAGUUGUUGGGGCCACUGAUUUUUCAUGCU- .(((((.((.((((...((.(((.(.((((..(((...(((((.....)))))..)))...)))).)(.(((------(....)))).)))).)))))).)).)))))...........- ( -36.80) >DroEre_CAF1 114152 116 + 1 UUAAG---AGUGAAAAUCAUUUUCGGGCCCCUGGCUGGCUGGCUAGUUGCCAAUCAGGUCACGAACUGUCGCUUCAGUCCAGUCAGUUUCAGUUUUCGGGGCCCCUGAUUUUUCAUGUU- .....---.((((((((((.....(((((((.((((((((((((.((.(((.....))).))(.((((......)))).))))))))..)))))...))))))).))).)))))))...- ( -43.50) >DroYak_CAF1 114434 112 + 1 UUAAG---AGUGAAAAUCAUUUUCGGGGCUCUGGCUUGCUGGCUAGUUGCCAAUCAGGUCACGAACUGUCGCUUCAGUCCA----GUUUCAGUUUUCGGGGCCCCUGAUUUUUCAUGUU- .....---.((((((((((.....((((((((((((((.((((.....))))..))))))..((((((..(((.......)----))..))))))..))))))))))).)))))))...- ( -40.20) >DroAna_CAF1 103069 109 + 1 UUAAG---AGUGAAAAUCAUUUUCGGCGCUUUUCCUUUCGGGCUAGUUGCCAAUCAGGUCACGAACUGUCGCUUCAGUCCG----GCGUUUGG--UCCGGG-CCCUGAUUUUUCAUGUU- .....---.((((((.........((((((..(((....)))..)).))))(((((((...(((....))).....(((((----((.....)--.)))))-))))))))))))))...- ( -32.10) >DroPer_CAF1 134480 113 + 1 UUCAGUAGCGCCAAAAUCAUCUGCAGUCCUUCUUUUUUUCGGCUGCUUGCCAAUCAGGUCACGGAGUGUCGC------CCGGUGGCGUCCAGUUGUUGGGGCCACUGAUUUUUCAUGCU- .(((((.((.((((...(....((((((............))))))..((((.((.(((.((.....)).))------).)))))).....)...)))).)).)))))...........- ( -33.80) >consensus UUAAG___AGUGAAAAUCAUUUUCGGGCCUCUGGCUUGCUGGCUAGUUGCCAAUCAGGUCACGAACUGUCGCUUCAGUCCA____GUUUCAGUUUUCGGGGCCCCUGAUUUUUCAUGUU_ .........(((((((......(((((.............(((.....))).....((((.(((((((.....................))))..))).))))))))).))))))).... (-13.94 = -15.13 + 1.20)

| Location | 17,004,511 – 17,004,624 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -33.79 |
| Consensus MFE | -14.76 |
| Energy contribution | -16.07 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17004511 113 - 22407834 AAACAUGAAAAAUCAGGGGCCCCGAAAACUGAAAC----UGGACUGAAGCGACAGUUCGUGACCUGAUUGGCAACUAGCCAGCAAGCCAGAGCCCCGAAAAUGAUUUUCACU---CUUAA .....((((((.((((((((...............----.((((((......)))))).((.(.((.(((((.....))))))).).))..))))).....)))))))))..---..... ( -28.30) >DroPse_CAF1 124906 113 - 1 -AGCAUGAAAAAUCAGUGGCCCCAACAACUGGACGCCACCGG------GCGACACUCCGUGACCUGAUUGGCAAGCAGCCAAAAAAAGAGCGGACUUCAGAUGAUUUUGGCGCUACUGAA -...........((((((((.((((((.(((((((((....)------)))....(((((.......(((((.....))))).......))))).))))).))...)))).)))))))). ( -41.14) >DroEre_CAF1 114152 116 - 1 -AACAUGAAAAAUCAGGGGCCCCGAAAACUGAAACUGACUGGACUGAAGCGACAGUUCGUGACCUGAUUGGCAACUAGCCAGCCAGCCAGGGGCCCGAAAAUGAUUUUCACU---CUUAA -....((((((.(((.(((((((.................((((((......)))))).....(((..((((.....))))..)))...))))))).....)))))))))..---..... ( -38.70) >DroYak_CAF1 114434 112 - 1 -AACAUGAAAAAUCAGGGGCCCCGAAAACUGAAAC----UGGACUGAAGCGACAGUUCGUGACCUGAUUGGCAACUAGCCAGCAAGCCAGAGCCCCGAAAAUGAUUUUCACU---CUUAA -....((((((.((((((((...............----.((((((......)))))).((.(.((.(((((.....))))))).).))..))))).....)))))))))..---..... ( -28.30) >DroAna_CAF1 103069 109 - 1 -AACAUGAAAAAUCAGGG-CCCGGA--CCAAACGC----CGGACUGAAGCGACAGUUCGUGACCUGAUUGGCAACUAGCCCGAAAGGAAAAGCGCCGAAAAUGAUUUUCACU---CUUAA -....((((((.(((((.-.((((.--.......)----)))......((((....))))..)))))(((((..((...((....))...)).)))))......))))))..---..... ( -29.50) >DroPer_CAF1 134480 113 - 1 -AGCAUGAAAAAUCAGUGGCCCCAACAACUGGACGCCACCGG------GCGACACUCCGUGACCUGAUUGGCAAGCAGCCGAAAAAAAGAAGGACUGCAGAUGAUUUUGGCGCUACUGAA -...........((((((((.((((.........((((.(((------(..((.....))..))))..))))..((((((...........)).))))........)))).)))))))). ( -36.80) >consensus _AACAUGAAAAAUCAGGGGCCCCGAAAACUGAAAC____CGGACUGAAGCGACAGUUCGUGACCUGAUUGGCAACUAGCCAGAAAGCCAGAGCCCCGAAAAUGAUUUUCACU___CUUAA .....((((((.((((((....)................(((((((......)))))))...)))))(((((.....)))))......................)))))).......... (-14.76 = -16.07 + 1.31)
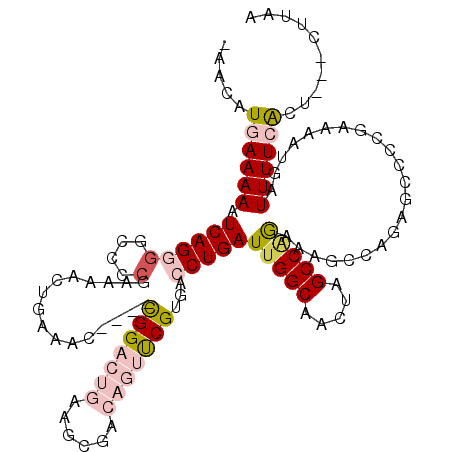
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:25 2006