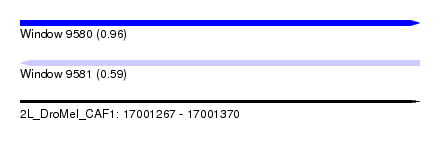
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,001,267 – 17,001,370 |
| Length | 103 |
| Max. P | 0.959850 |

| Location | 17,001,267 – 17,001,370 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
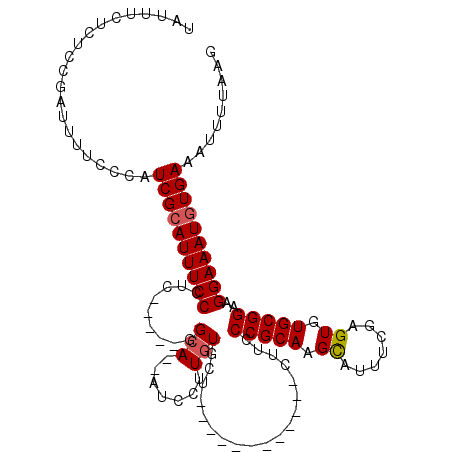
>2L_DroMel_CAF1 17001267 103 + 22407834 CAUUUCUCUCCGAUUUUCCCAUCGCAUUUCCCAC-----GGCA--AUCCUUGUGCU------UCUCUGCUUCCCGCAAGCAUUUCGAGUGUGCGGAAGGAAAUGUGAAAUUUUAAG .....................((((((((((..(-----.(((--.((...(((((------....(((.....))))))))...))...))).)..))))))))))......... ( -27.40) >DroSec_CAF1 108414 101 + 1 AAUUUCUCUCCGAUUUUCCCAUCGCAUUUCCCUC-----GGCA--AUCCUUGUGCUUGUG--------CUUCCCGCAAGCAUUUCGAGUGUGCGGAAGGAAAUGUGAAAUUUUAAG .....................((((((((((.((-----.(((--.((...(((((((((--------.....)))))))))...))...))).)).))))))))))......... ( -35.30) >DroSim_CAF1 110398 109 + 1 UAUUUCUCUCCGAUUUUCCCAUCGCAUUUCCCUC-----GGCA--AUCCUUGUGCUUGUGCUUCUGUGCUUCCCGCAAGCAUUUCGAGUGUGCGGAAGGAAAUGUGAAAUUUUAAG .....................((((((((((.((-----.(((--.((...((((((((((......)).....))))))))...))...))).)).))))))))))......... ( -34.30) >DroEre_CAF1 111040 95 + 1 UAUUUCUCUGCGAUUUUCCCAUCGCAUUUCCCUC-----GGCA--AUACUUGUG--------------CUUCCCGCAAGCAUUUCGAGUGUGCGGAAGGAAAUGUGAAAAUUUAAG .....................((((((((((.((-----.(((--.((((((((--------------(((.....)))))...))))))))).)).))))))))))......... ( -30.70) >DroYak_CAF1 111211 95 + 1 UAUUUCUCUCCGAUUUUCCCAUCGCAUUUCCCUC-----GGCA--AUUCUUGUG--------------CUUCCCGCACGCUUUUCGAGUGUGCGGAAGGAAAUGUGAAAUUUUAAG .....................((((((((((...-----((((--.......))--------------))..((((((((((...))))))))))..))))))))))......... ( -31.80) >DroAna_CAF1 99963 111 + 1 GAUCCUGGCCAGAUUCUCCACUCG-AUUUCCCCCUAUCCCGCACCACCUUUAUCCGUU----CGUUGGGUUCCCGCAAGUGGUGUGAGUGUGCGGCAGGAAAUGUGAAAUUUUAAG ..(((((.(((.((((.......(-((........)))..(((((((....(((((..----...)))))........))))))))))).)).).)))))................ ( -28.70) >consensus UAUUUCUCUCCGAUUUUCCCAUCGCAUUUCCCUC_____GGCA__AUCCUUGUGCU____________CUUCCCGCAAGCAUUUCGAGUGUGCGGAAGGAAAUGUGAAAUUUUAAG .....................((((((((((.........(((.......)))...................(((((.((.......)).)))))..))))))))))......... (-19.62 = -19.82 + 0.20)

| Location | 17,001,267 – 17,001,370 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17001267 103 - 22407834 CUUAAAAUUUCACAUUUCCUUCCGCACACUCGAAAUGCUUGCGGGAAGCAGAGA------AGCACAAGGAU--UGCC-----GUGGGAAAUGCGAUGGGAAAAUCGGAGAGAAAUG ......(((((.((((((((((((((..(.......)..))))))).((.....------.)).((.((..--..))-----.)))))))))((((......))))....))))). ( -26.40) >DroSec_CAF1 108414 101 - 1 CUUAAAAUUUCACAUUUCCUUCCGCACACUCGAAAUGCUUGCGGGAAG--------CACAAGCACAAGGAU--UGCC-----GAGGGAAAUGCGAUGGGAAAAUCGGAGAGAAAUU .....((((((.((((((((((.(((.(..(....((((((.(.....--------).))))))...)..)--))).-----))))))))))((((......))))....)))))) ( -31.00) >DroSim_CAF1 110398 109 - 1 CUUAAAAUUUCACAUUUCCUUCCGCACACUCGAAAUGCUUGCGGGAAGCACAGAAGCACAAGCACAAGGAU--UGCC-----GAGGGAAAUGCGAUGGGAAAAUCGGAGAGAAAUA ......(((((.((((((((((.(((.(..(....((((((.(.............).))))))...)..)--))).-----))))))))))((((......))))....))))). ( -29.62) >DroEre_CAF1 111040 95 - 1 CUUAAAUUUUCACAUUUCCUUCCGCACACUCGAAAUGCUUGCGGGAAG--------------CACAAGUAU--UGCC-----GAGGGAAAUGCGAUGGGAAAAUCGCAGAGAAAUA ......(((((..(((((((((.(((........(((((((.(.....--------------).)))))))--))).-----)))))))))(((((......))))).)))))... ( -29.60) >DroYak_CAF1 111211 95 - 1 CUUAAAAUUUCACAUUUCCUUCCGCACACUCGAAAAGCGUGCGGGAAG--------------CACAAGAAU--UGCC-----GAGGGAAAUGCGAUGGGAAAAUCGGAGAGAAAUA ......(((((.(((((((((((((((.((.....)).)))))....(--------------((.......--))).-----))))))))))((((......))))....))))). ( -29.30) >DroAna_CAF1 99963 111 - 1 CUUAAAAUUUCACAUUUCCUGCCGCACACUCACACCACUUGCGGGAACCCAACG----AACGGAUAAAGGUGGUGCGGGAUAGGGGGAAAU-CGAGUGGAGAAUCUGGCCAGGAUC .......(((((((((((((.((.....(((.((((((((..((....))..(.----...).....)))))))).)))...)))))))))-...)))))).((((.....)))). ( -30.10) >consensus CUUAAAAUUUCACAUUUCCUUCCGCACACUCGAAAUGCUUGCGGGAAG____________AGCACAAGGAU__UGCC_____GAGGGAAAUGCGAUGGGAAAAUCGGAGAGAAAUA ......(((((.((((((((((((((.............))))))).((............))......................)))))))((((......))))....))))). (-15.82 = -16.52 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:20 2006