| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
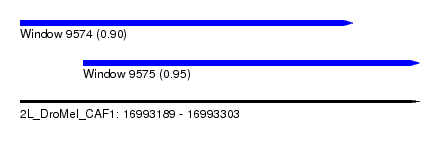
| Location | 16,993,189 – 16,993,303 |
| Length | 114 |
| Max. P | 0.951005 |

| Location | 16,993,189 – 16,993,284 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -15.62 |
| Consensus MFE | -12.90 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16993189 95 + 22407834 CUCAACUGCCCUUAAUUCCUUA--CUUAAUUCCACACAAUUUCUGGACCAAGUCGUGCGCUUAACAACUUGUCCAGGAGCCCACAAGAUCCCCGAAC ......................--...............(((((((((.((((.((.......)).))))))))))))).................. ( -15.90) >DroSec_CAF1 100448 95 + 1 CUCAACUGCCCUUAAUUCCUUA--CUUAAUUCCACACAAUUUCUGGACCAAGUCGUGCGCUUAACAACUUGUCCAGGAGCCCACAAGAUCCCCGAAC ......................--...............(((((((((.((((.((.......)).))))))))))))).................. ( -15.90) >DroSim_CAF1 102403 95 + 1 CUCAACUGCCCUUAAUUCCUUA--CUUAAUUCCACACAAUUUCUGGACCAAGUCGUGCGCUUAACAACUUGUCCGGGAGCCCACAAAAUCCCCGAAC ......................--...............(((((((((.((((.((.......)).))))))))))))).................. ( -15.20) >DroEre_CAF1 103194 94 + 1 CACAACUGCCCUUAAUACCUUA--CUUAAUUCCACACAAUUUCUGGACCAAGU-GUGUGCCUAACCACUUGUCCAGGAGCCCACAAGAUCCCCGAAC ......................--...............(((((((((.((((-(.((.....)))))))))))))))).................. ( -18.90) >DroYak_CAF1 103201 85 + 1 CUCAACUGCGCUUAAUUCCUUG--CUUAAUUUCACACAAUUUCUGGACCAAGUAGUGCACCUAACAACUUGUCCAGAAGUCCACA----------AC .........((..........)--).............((((((((((.((((.((.......)).)))))))))))))).....----------.. ( -17.80) >DroAna_CAF1 92181 93 + 1 ----ACAGCCUGUAAUUCUUCCCACACGAUUUCCUCAAACUCCUGGCUCAGUUUAAGCGCUUUACAACUUGUCCCAGAGCCCACAAGAUCCCUGAGC ----.............................((((.....(((((((.(..((((..........))))..)..)))))....)).....)))). ( -10.00) >consensus CUCAACUGCCCUUAAUUCCUUA__CUUAAUUCCACACAAUUUCUGGACCAAGUCGUGCGCUUAACAACUUGUCCAGGAGCCCACAAGAUCCCCGAAC .......................................((((((((.(((((.((.......)).))))))))))))).................. (-12.90 = -13.23 + 0.34)


| Location | 16,993,207 – 16,993,303 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.70 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.35 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16993207 96 + 22407834 CUUACUUAAUUCCACACAAUUUCUGG-ACCAAGUCGUGCGCUUAACAACUUGUCCAGGAGCCCACAAGAUCCCCGAACUCCCAGCUCCUGGCCAG----U---------C ......................((((-..(((((.((.......)).))))).((((((((.....((.((...)).))....))))))))))))----.---------. ( -23.00) >DroPse_CAF1 110984 98 + 1 CCUACUAUUUUCCAU-CAAUUUGUGGGGCCGGCUUAAGCGCUCAACAACUUGUCCGAGAU-----------CGGGUACUUGCUGCUGCCUGCCAGAUUCCCAGGACAAUC ...............-....((((.((((..((....)))))).)))).((((((..(((-----------(.((((...((....)).)))).))))....)))))).. ( -24.10) >DroSec_CAF1 100466 96 + 1 CUUACUUAAUUCCACACAAUUUCUGG-ACCAAGUCGUGCGCUUAACAACUUGUCCAGGAGCCCACAAGAUCCCCGAACUACCAGCUCCUGGCCAG----U---------C ......................((((-..(((((.((.......)).))))).((((((((.....((.((...)).))....))))))))))))----.---------. ( -23.00) >DroSim_CAF1 102421 96 + 1 CUUACUUAAUUCCACACAAUUUCUGG-ACCAAGUCGUGCGCUUAACAACUUGUCCGGGAGCCCACAAAAUCCCCGAACUCCCAGCUUCUGGCCAG----U---------A ......................((((-..(((((.((.......)).))))).((((((((......................))))))))))))----.---------. ( -18.95) >DroEre_CAF1 103212 94 + 1 CUUACUUAAUUCCACACAAUUUCUGG-ACCAAGU-GUGUGCCUAACCACUUGUCCAGGAGCCCACAAGAUCCCCGAACUCCCAGCUCCUGGCUA-----U---------C ..........((((.........)))-).(((((-(.((.....)))))))).((((((((.....((.((...)).))....))))))))...-----.---------. ( -22.90) >DroYak_CAF1 103219 90 + 1 CUUGCUUAAUUUCACACAAUUUCUGG-ACCAAGUAGUGCACCUAACAACUUGUCCAGAAGUCCACA----------ACUUCCAGCUCCUGGCUC-----CAAGGA----G ..................((((((((-((.((((.((.......)).)))))))))))))).....----------........((((((....-----).))))----) ( -23.10) >consensus CUUACUUAAUUCCACACAAUUUCUGG_ACCAAGUCGUGCGCUUAACAACUUGUCCAGGAGCCCACAAGAUCCCCGAACUCCCAGCUCCUGGCCAG____U_________C .......................(((...(((((.((.......)).))))).((((((((......................)))))))))))................ (-10.73 = -11.35 + 0.62)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:15 2006