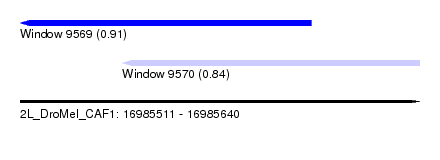
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,985,511 – 16,985,640 |
| Length | 129 |
| Max. P | 0.907848 |

| Location | 16,985,511 – 16,985,605 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16985511 94 - 22407834 CUAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAACAAUGAGGACAUGCAAAUGCAGUUUAAGCUCAA .......((((((...(((....)))..)))))).((..((((((....(((((((......)))).)))..(((....))))))))))).... ( -21.60) >DroSec_CAF1 93151 94 - 1 CUAUACCCGGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAACAAUGAGGACAUGCAAAUGCAGUUUAAGCUCAA .........(((....))).((((((....((((((.(((.............))))))))).....((((.(((....))))))).)))))). ( -20.92) >DroSim_CAF1 95113 94 - 1 CUAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAACAAUGAGGACAUGCAAAUGCAGUUUAAGCUCAA .......((((((...(((....)))..)))))).((..((((((....(((((((......)))).)))..(((....))))))))))).... ( -21.60) >DroEre_CAF1 95789 93 - 1 CCAUUCCCAGCUGUUAAGCGUGCGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAACAAAGAGGAAAUGCAAAUGCAGUUU-AGCUCAA ............(((((((.(((((........)))))..........((((...((........)))))).(((....)))))))-))).... ( -17.60) >DroYak_CAF1 95711 94 - 1 CCAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAACAAAGAGGACAUGCAAAUGCAGUUUAAGCUCAA .........(((....))).((((((....((((((.(((.............))))))))).....((((.(((....))))))).)))))). ( -21.12) >consensus CUAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAACAAUGAGGACAUGCAAAUGCAGUUUAAGCUCAA .......((((((...(((....)))..)))))).((............(((((((......)))).)))..(((....)))......)).... (-19.24 = -19.48 + 0.24)

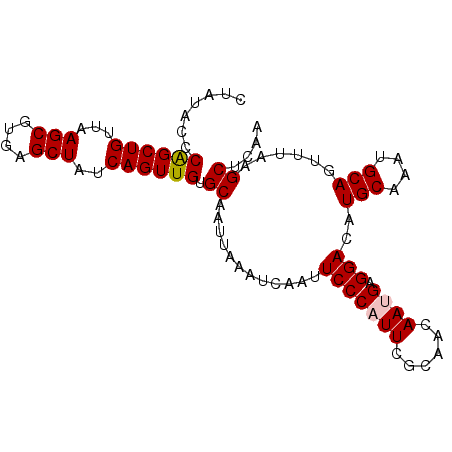

| Location | 16,985,544 – 16,985,640 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.48 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16985544 96 - 22407834 UGAUGCUUAAUUGGGUUUUAGUUUUCGGCCAAAUCCUAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAA ((((..((((((((((..(((..((......))..))).)))((((((...(((....)))..))))))..))))))))))).............. ( -22.20) >DroSec_CAF1 93184 96 - 1 UGAUGCUUAAUUGGGUUUUAGUUUUCGGCCAAAUUCUAUACCCGGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAA ...(((.....((((..((.((((...((.............((((((...(((....)))..)))))).))....)))).))..))))...))). ( -21.24) >DroSim_CAF1 95146 96 - 1 UGAUGCUUAAUUGGGUUUUAGUUUUCGGCCAAAUUCUAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAA ...(((.....((((..((.((((...((.............((((((...(((....)))..)))))).))....)))).))..))))...))). ( -22.04) >DroEre_CAF1 95821 96 - 1 UGAUGCUUAAUUGGGUUUUAGUUUUCAGCCAAAUUCCAUUCCCAGCUGUUAAGCGUGCGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAA ...(((.....((((..((.((((.((((...............))))..)))).(((((........)))))........))..))))...))). ( -20.16) >DroYak_CAF1 95744 96 - 1 UGAUGUUUAAUUGGGUUUUAGUUUUCAGCCAAAUUCCAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAA ....(((((((((((((.........))))............((((((...(((....)))..))))))..)))))))))................ ( -21.10) >consensus UGAUGCUUAAUUGGGUUUUAGUUUUCGGCCAAAUUCUAUACCCAGCUGUUAAGCGUGAGCUAUCAGUUGUGCAAUUAAAUCAAUUCCCAUUCGCAA ...(((.....((((..((.((((...((.............((((((...(((....)))..)))))).))....)))).))..))))...))). (-20.80 = -20.48 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:10 2006