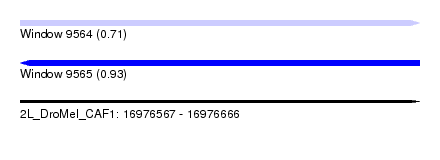
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,976,567 – 16,976,666 |
| Length | 99 |
| Max. P | 0.925431 |

| Location | 16,976,567 – 16,976,666 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.46 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -16.97 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16976567 99 + 22407834 ACUUAUUUUCCGGAUG-------AUCAAGGGGA--AAAAUGUGCCGAGAUUGAUACACAAUGUUUGCAUUGUGUAACCAAUCCGCUUGAGUGGGUUACUAU------UCUCGUC .....((..((.....-------.....))..)--)........(((((....(((((((((....)))))))))...(((((((....))))))).....------))))).. ( -28.70) >DroSec_CAF1 84357 99 + 1 ACUUAUUUUCCGGAUG-------AUCAAGGGGA--AAAAAGUGCCGAGAUUGAUACACAAUGUUUGCAUUGUGUAACCAAUCCGCUUGAGUGAGUUACUAU------CCUCGUC ((((((((..((((((-------(((..((.(.--......).))..))))..(((((((((....)))))))))....)))))...))))))))......------....... ( -27.10) >DroSim_CAF1 86111 85 + 1 --------------UG-------AUCAAGGGGA--AAAAAGUGCCGAGAUUGAUACACAAUGUUUGCAUUGUGUAACCAAUCCGCUUGAGUGAGUUACUAU------CCUCGUC --------------..-------.....(((((--...(((((....(((((.(((((((((....)))))))))..)))))))))).((((...)))).)------))))... ( -24.80) >DroEre_CAF1 86732 99 + 1 ACUUAUUUUCCGAAUG-------AUCAAGGGGA--AACAAGUGCCGAGAUUGAUACACAAUGUUUGCAUUGUGUAACCAAUCCGCUUGAGUGAGUUAGUAU------CCUCGUC (((((((..((.....-------.....))..)--..((((((....(((((.(((((((((....)))))))))..))))))))))).))))))......------....... ( -27.00) >DroYak_CAF1 85924 99 + 1 ACUUAUUUCCCGAAUG-------AUCAAGGGGA--AACAAGUGCCGAGAUUGAUACACAAAGUUUGCAUUGUGUAACCAAUCCGCUUCAGUGAGUUGGUAU------CCUCGUA ((((.((((((.....-------......))))--)).))))..((((.....(((((((.(....).)))))))(((((..(((....)))..)))))..------.)))).. ( -30.60) >DroAna_CAF1 78128 114 + 1 UCUGAUAUCCCAGCUGCCAGAAUGUCGGGGAGAGAAAGUAGCUCUGAGAUUGAGAAAUACUGUUUGCAUUGUGUAACUAAUCCAGUUGGGUAAGCUACUACUGCUGCCUUUGUC ...((((.((((((((.((((((.((((((..(.....)..)))))).))).(....).))).((((.....))))......))))))))..(((.......))).....)))) ( -23.80) >consensus ACUUAUUUUCCGGAUG_______AUCAAGGGGA__AAAAAGUGCCGAGAUUGAUACACAAUGUUUGCAUUGUGUAACCAAUCCGCUUGAGUGAGUUACUAU______CCUCGUC ............................((((............((.(((((.(((((((((....)))))))))..)))))))(((....))).............))))... (-16.97 = -16.42 + -0.55)


| Location | 16,976,567 – 16,976,666 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.46 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16976567 99 - 22407834 GACGAGA------AUAGUAACCCACUCAAGCGGAUUGGUUACACAAUGCAAACAUUGUGUAUCAAUCUCGGCACAUUUU--UCCCCUUGAU-------CAUCCGGAAAAUAAGU ..(((((------........((.(....).)).(((((.((((((((....))))))))))))))))))......(((--(((.......-------.....))))))..... ( -23.20) >DroSec_CAF1 84357 99 - 1 GACGAGG------AUAGUAACUCACUCAAGCGGAUUGGUUACACAAUGCAAACAUUGUGUAUCAAUCUCGGCACUUUUU--UCCCCUUGAU-------CAUCCGGAAAAUAAGU ...(((.------.......)))......((((((((((.((((((((....))))))))))))))))..))(((((((--(((.......-------.....)))))).)))) ( -27.20) >DroSim_CAF1 86111 85 - 1 GACGAGG------AUAGUAACUCACUCAAGCGGAUUGGUUACACAAUGCAAACAUUGUGUAUCAAUCUCGGCACUUUUU--UCCCCUUGAU-------CA-------------- (((((((------..(((.....)))...((((((((((.((((((((....))))))))))))))))..)).......--...))))).)-------).-------------- ( -25.40) >DroEre_CAF1 86732 99 - 1 GACGAGG------AUACUAACUCACUCAAGCGGAUUGGUUACACAAUGCAAACAUUGUGUAUCAAUCUCGGCACUUGUU--UCCCCUUGAU-------CAUUCGGAAAAUAAGU ...(((.------.......)))......((((((((((.((((((((....))))))))))))))))..))(((((((--(.((...((.-------...)))).)))))))) ( -28.60) >DroYak_CAF1 85924 99 - 1 UACGAGG------AUACCAACUCACUGAAGCGGAUUGGUUACACAAUGCAAACUUUGUGUAUCAAUCUCGGCACUUGUU--UCCCCUUGAU-------CAUUCGGGAAAUAAGU ...(((.------.......)))......((((((((((.((((((........))))))))))))))..))(((((((--((((...((.-------...))))))))))))) ( -32.20) >DroAna_CAF1 78128 114 - 1 GACAAAGGCAGCAGUAGUAGCUUACCCAACUGGAUUAGUUACACAAUGCAAACAGUAUUUCUCAAUCUCAGAGCUACUUUCUCUCCCCGACAUUCUGGCAGCUGGGAUAUCAGA .......(((...((.((((((...((....))...))))))))..)))...............(((((((.((((.....((.....)).....))))..)))))))...... ( -22.50) >consensus GACGAGG______AUAGUAACUCACUCAAGCGGAUUGGUUACACAAUGCAAACAUUGUGUAUCAAUCUCGGCACUUUUU__UCCCCUUGAU_______CAUCCGGAAAAUAAGU ..............................(((((((((.((((((((....))))))))))))))).))............................................ (-14.19 = -14.92 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:05 2006