
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,967,761 – 16,967,902 |
| Length | 141 |
| Max. P | 0.995605 |

| Location | 16,967,761 – 16,967,871 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -14.82 |
| Energy contribution | -14.18 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16967761 110 - 22407834 CAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC----------UAUCUUCGGCUUUCACUCGCCUCUCCUAGUUUUUACACUGUGAAGAAAAUAUAUUUUACAAACGGAAAAUAAAACA .(((((((((............(((........)----------)).....(((........))).)))))))))......((((....((((....))))....))))........... ( -19.10) >DroSec_CAF1 75457 101 - 1 CAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC----------UAUCUUCGGCUUUCACUCGCCUCUUCUAGUUUCUACACUGUGAAGAAAAUAUAUAUUACAAAC---------AACA ..((((((((........))))))))........----------.......(((........)))(((((((((......)))).))))).................---------.... ( -16.50) >DroSim_CAF1 77529 98 - 1 CAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC----------UAUCUUCGGCUUUCACUCGCCUCUUCUAGUUUCUACACCGUGAAGAAAAUAUAUAUUACAAAC---------G--- ..((((((((........))))))))........----------..((((((((........))).......((....)).....))))).................---------.--- ( -14.80) >DroEre_CAF1 78095 110 - 1 CAGCUAGGAGUUCUCCGCUUGCUGGUCCUCCCUCUUCUCCUCCCUAUCUUCGGCUUUCACUCGCCUCUUCUAGUUCUUACACAGCGAAGAAAAUACUU--UCCUAUCAGCCA-------- ((((..(((....)))....))))......................((((((((........))).......((....)).....)))))........--............-------- ( -18.10) >DroYak_CAF1 77059 111 - 1 CAGCUAGGAGUUCUCCGCUUUCUAGUCCCCCCUC------UCUCUAACUUCGGCUUUCACUCGCCUCUCCUAGUUCUUACACCGCGAAUAAAAUAUUU--GCCCAU-AGCCAUUGAAAGC .(((((((((......(((....)))........------...........(((........))).)))))))))........(((((((...)))))--))....-............. ( -20.40) >consensus CAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC__________UAUCUUCGGCUUUCACUCGCCUCUUCUAGUUUUUACACCGUGAAGAAAAUAUAU_UUACAAAC_G__A____AA__ .(((((((((......(((....))).........................(((........))).)))))))))............................................. (-14.82 = -14.18 + -0.64)

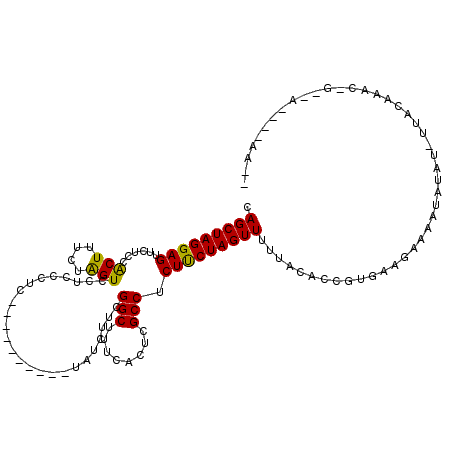
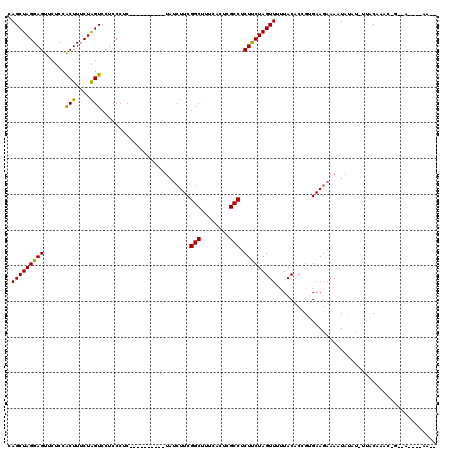
| Location | 16,967,801 – 16,967,902 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16967801 101 + 22407834 GUAAAAACUAGGAGAGGCGAGUGAAAGCCGAAGAUA----------GAGGGAGGACUAGAAAGUGGAGAACUCCUAGCUGUUGGCUCAACUUCAAAUUGCCAGCGGGGCAU ((.....(((((((.(((........))).......----------........(((....)))......)))))))((((((((.............)))))))).)).. ( -28.52) >DroSec_CAF1 75488 101 + 1 GUAGAAACUAGAAGAGGCGAGUGAAAGCCGAAGAUA----------GAGGGAGGACUAGAAAGUGGAGAACUCCUAGCUGUUGGCUCAACUUCAAAUUGCCAGCGGGGCAU .......(((.....(((........))).....))----------).(((((..((.........))..)))))..((((((((.............))))))))..... ( -26.62) >DroSim_CAF1 77557 101 + 1 GUAGAAACUAGAAGAGGCGAGUGAAAGCCGAAGAUA----------GAGGGAGGACUAGAAAGUGGAGAACUCCUAGCUGUUGGCUCAACUUCAAAUUGCCAGCGGGGCAU .......(((.....(((........))).....))----------).(((((..((.........))..)))))..((((((((.............))))))))..... ( -26.62) >DroEre_CAF1 78125 111 + 1 GUAAGAACUAGAAGAGGCGAGUGAAAGCCGAAGAUAGGGAGGAGAAGAGGGAGGACCAGCAAGCGGAGAACUCCUAGCUGUUGGCUCAACUUCAAAUUGCCAGCGGGGCAU .......(((.....(((........))).....))).........((((..(..((((((.(((((....)))..))))))))..)..))))....((((.....)))). ( -32.00) >DroYak_CAF1 77096 105 + 1 GUAAGAACUAGGAGAGGCGAGUGAAAGCCGAAGUUAGAGA------GAGGGGGGACUAGAAAGCGGAGAACUCCUAGCUGUUGGCUCAACUUCAAAUUGCCAGCGGGGCAU ((.....(((((((.(((........))).......(...------...((....))......)......)))))))((((((((.............)))))))).)).. ( -31.32) >consensus GUAAAAACUAGAAGAGGCGAGUGAAAGCCGAAGAUA__________GAGGGAGGACUAGAAAGUGGAGAACUCCUAGCUGUUGGCUCAACUUCAAAUUGCCAGCGGGGCAU ..........................(((...................(((((..((.........))..)))))..((((((((.............))))))))))).. (-24.26 = -24.30 + 0.04)

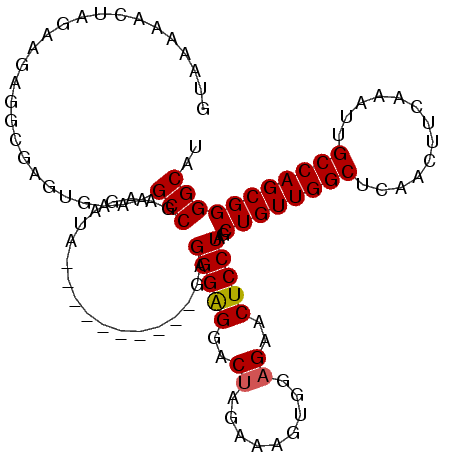
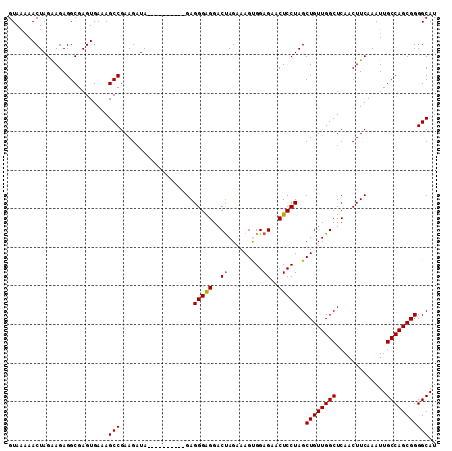
| Location | 16,967,801 – 16,967,902 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16967801 101 - 22407834 AUGCCCCGCUGGCAAUUUGAAGUUGAGCCAACAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC----------UAUCUUCGGCUUUCACUCGCCUCUCCUAGUUUUUAC .((((.....)))).......((((...))))(((((((((............(((........)----------)).....(((........))).)))))))))..... ( -21.90) >DroSec_CAF1 75488 101 - 1 AUGCCCCGCUGGCAAUUUGAAGUUGAGCCAACAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC----------UAUCUUCGGCUUUCACUCGCCUCUUCUAGUUUCUAC .((((.....))))....((((..(((......((((((((........)))))))).....)))----------...))))(((........)))............... ( -21.60) >DroSim_CAF1 77557 101 - 1 AUGCCCCGCUGGCAAUUUGAAGUUGAGCCAACAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC----------UAUCUUCGGCUUUCACUCGCCUCUUCUAGUUUCUAC .((((.....))))....((((..(((......((((((((........)))))))).....)))----------...))))(((........)))............... ( -21.60) >DroEre_CAF1 78125 111 - 1 AUGCCCCGCUGGCAAUUUGAAGUUGAGCCAACAGCUAGGAGUUCUCCGCUUGCUGGUCCUCCCUCUUCUCCUCCCUAUCUUCGGCUUUCACUCGCCUCUUCUAGUUCUUAC .((((.....))))....((((..(((....((((..(((....)))....))))...)))...))))..............(((........)))............... ( -24.20) >DroYak_CAF1 77096 105 - 1 AUGCCCCGCUGGCAAUUUGAAGUUGAGCCAACAGCUAGGAGUUCUCCGCUUUCUAGUCCCCCCUC------UCUCUAACUUCGGCUUUCACUCGCCUCUCCUAGUUCUUAC .((((.....))))....(((((((((......((((((((........))))))))........------.))).))))))(((........)))............... ( -22.56) >consensus AUGCCCCGCUGGCAAUUUGAAGUUGAGCCAACAGCUAGGAGUUCUCCACUUUCUAGUCCUCCCUC__________UAUCUUCGGCUUUCACUCGCCUCUUCUAGUUUUUAC .((((.....)))).......((((...))))(((((((((......(((....))).........................(((........))).)))))))))..... (-20.82 = -20.18 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:03 2006