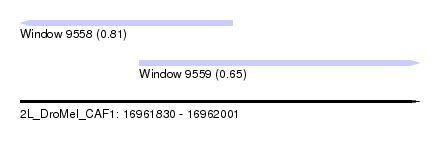
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,961,830 – 16,962,001 |
| Length | 171 |
| Max. P | 0.812542 |

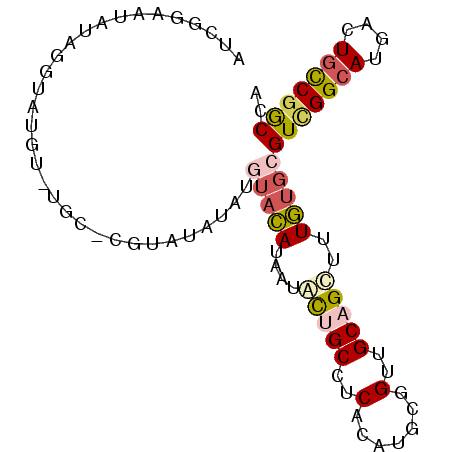
| Location | 16,961,830 – 16,961,921 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -15.62 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16961830 91 - 22407834 AUCGGAAUAUAGGUAUGUAUGCUCGUAUAUAUGUACAUAAUGCUGCCUCACAUGCGGUUGCAGCUUUGUGCGUCGGCAUGACUGCCAACCA ...........((((..(((((.((.....(((((((...(((.(((.(....).))).)))....))))))))))))))..))))..... ( -26.20) >DroSec_CAF1 69476 91 - 1 AUCGGAAUAUAGGUAUGUAUGCACGUAUAUAUGUACAUAAUGCUGCCUCACAUGCGGUUGCAGCUUUGUGCGUCGGCAUGACUGCCGGCCA ............((((((((....))))))))(((((...(((.(((.(....).))).)))....)))))(((((((....))))))).. ( -29.70) >DroSim_CAF1 71637 91 - 1 AUCGGAAUAUAGGUAUGUAUGCACGUAUAUAUGUACAUAAUGCUGCCUCACAUGCGGUUGCAGCUUUGUGCGUCGGCAUGACUGCCGGCCA ............((((((((....))))))))(((((...(((.(((.(....).))).)))....)))))(((((((....))))))).. ( -29.70) >DroEre_CAF1 72288 83 - 1 AUCGGAAUAUAUGUAUGU--------UUGUAUGUACAUAAUAUUGCCUCACAUGCGGUUGCAGCUUUGUGCGUCGGCAUGGCUGCCGGCCA ...(((((((((((((((--------....)))))))).))))).))........((((((((((..((((....))))))))).))))). ( -27.80) >DroYak_CAF1 71146 83 - 1 AUAGAAAUAUAUGUACGU--------UUGUAUAUACAUAAUAUGGCCUCACAUGCGGUUGCAGCUUUGUGCGUCGGCAUGACUGUCGGCCA .........(((((((..--------..))))))).......(((((.(((((((.(.((((......)))).).)))))..))..))))) ( -23.80) >DroAna_CAF1 63785 72 - 1 -------------------CACUCGCACACACAACUACCAGACAGCCUCACAUGCGGUUGCAGUUUUAUGCGUUGGCAUGACUCCCGGCCA -------------------.........................(((...(((((.(.((((......)))).).)))))......))).. ( -14.40) >consensus AUCGGAAUAUAGGUAUGU_UGC_CGUAUAUAUGUACAUAAUACUGCCUCACAUGCGGUUGCAGCUUUGUGCGUCGGCAUGACUGCCGGCCA ................................(((((....(((((..(.......)..)))))..)))))(((((((....))))))).. (-15.62 = -16.23 + 0.62)

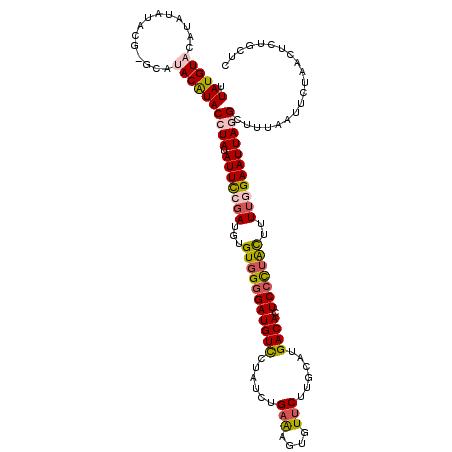
| Location | 16,961,881 – 16,962,001 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -17.56 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16961881 120 + 22407834 UUAUGUACAUAUAUACGAGCAUACAUACCUAUAUUCCGAUGUGUUGCGAUGUCCUAUCUGAAAGUGUUCUUGCACGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCUUCUC .(((((.(........).)))))....((((.(((((((...((.(.(((((((.....)...((((....)))))))).)).).))..))))))))))).................... ( -20.70) >DroSec_CAF1 69527 120 + 1 UUAUGUACAUAUAUACGUGCAUACAUACCUAUAUUCCGAUGUGUGGGGAUGUCCUAUCUGAAAGUGUUCUUGCAUGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCUGCUC .(((((((........)))))))....((((.(((((((...(((((((((((......(((....)))......)))).)))))))..))))))))))).................... ( -34.10) >DroSim_CAF1 71688 120 + 1 UUAUGUACAUAUAUACGUGCAUACAUACCUAUAUUCCGAUGUGUGGGGAUGUCCUAUCUGAAAGUGUGCUUGCAUGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCUAUUC .(((((((........)))))))....((((.(((((((...(((((((((((.....((.(((....))).)).)))).)))))))..))))))))))).................... ( -34.00) >DroEre_CAF1 72339 112 + 1 UUAUGUACAUACAA--------ACAUACAUAUAUUCCGAUGUGUGGGGAUGUUCCAUUUGAGAGCGUUCUGGCAUGACACUCCUUGUUUUUUGAAUUAGGCUAAAACUCGAACUCUGCUC ((((((........--------.(((((((........)))))))((((((((((....).))))))))).))))))....(((.((((...)))).))).................... ( -21.80) >DroYak_CAF1 71197 112 + 1 UUAUGUAUAUACAA--------ACGUACAUAUAUUUCUAUGUGUGGGGAUGUUAUAUCUGAGGGUGUUCUGGCAAGACACUCCUUAUUUUUCGAAUUAGGCUAAAAUUCGAACUCUGCUC ....(((((.(((.--------.(.((((((((....)))))))).)..)))))))).(((((((((.((....)))))).)))))...((((((((.......))))))))........ ( -28.20) >consensus UUAUGUACAUAUAUACG_GCAUACAUACCUAUAUUCCGAUGUGUGGGGAUGUCCUAUCUGAAAGUGUUCUUGCAUGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCUGCUC .((((((..............))))))((((.(((((((...(((((((((((......(((....)))......)))).)))))))..))))))))))).................... (-17.56 = -18.12 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:00 2006