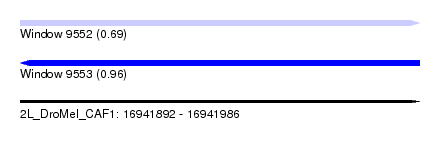
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,941,892 – 16,941,986 |
| Length | 94 |
| Max. P | 0.958853 |

| Location | 16,941,892 – 16,941,986 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -15.03 |
| Energy contribution | -15.03 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
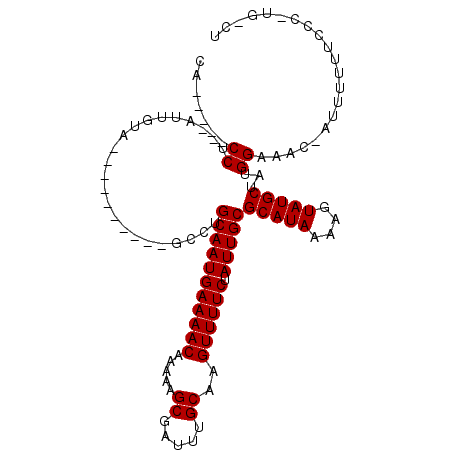
>2L_DroMel_CAF1 16941892 94 + 22407834 CG-----CCU--GUUUUA----------GCCUGCAAUGAAAACAAAAAGCGAUUUGCAAGUUUUCUAUUGCGCAUAAAAGUAUGCUAUGGAACCUAUUUUUCCCUCCGCUU ..-----...--.....(----------((..(((((((((((.....((.....))..)))))).)))))(((((....)))))...((((.......))))....))). ( -17.20) >DroVir_CAF1 69649 94 + 1 CAUUCGUCCU-----GUU----------GCCUGCAAUGAAAACAAAAAGCGAUUUGCAAGUUUUCUAUUGCGCAUAAAAGUAUGCUGUGGAAA--AUUUUUUCCCAUGACG ....((((.(-----(..----------....(((((((((((.....((.....))..)))))).)))))(((((....)))))...(((((--....))))))).)))) ( -21.10) >DroPse_CAF1 56412 98 + 1 CA-----CCC--AUUGGAUGGAUCCU--GGCUGCAAUGAAAACAAAAAGCGAUUUGCAAGUUUUCUAUUGCGCAUAAAAGUAUGCUAUGGAAGCCAUUUUUUCAC----UU ..-----.((--((...))))....(--(((((((((((((((.....((.....))..)))))).)))))(((((....)))))......))))).........----.. ( -22.80) >DroGri_CAF1 58357 99 + 1 CACCCCUCCUCACUCCUU----------GCCGGCAAUGAAAACAAAAAGCGAUUUGCAAGUUUUCUAUUGCGCAUAAAAGUAUGCUGUGGGAA--AUUUUUUCCCAUGACG ..................----------....(((((((((((.....((.....))..)))))).)))))(((((....))))).(((((((--.....))))))).... ( -22.60) >DroAna_CAF1 50652 99 + 1 UG-----CCU--GUUUUA----UCCUCGGCCUGCAAUGAAAACAAAAAGCGAUUUGCAAGUUUUCUAUUGCGCAUAAAAGUAUGCUAUGGAUCCUAUUUUU-CCCCUGCCA ..-----...--......----.....(((..(((((((((((.....((.....))..)))))).)))))(((((....)))))...(((.........)-))...))). ( -19.80) >DroPer_CAF1 57046 98 + 1 CA-----CCC--AUUGGAUGGAUCCU--GGCUGCAAUGAAAACAAAAAGCGAUUUGCAAGUUUUCUAUUGCGCAUAAAAGUAUGCUAUGGAAGCCAUUUUUUCAC----UU ..-----.((--((...))))....(--(((((((((((((((.....((.....))..)))))).)))))(((((....)))))......))))).........----.. ( -22.80) >consensus CA_____CCU__AUUGUA__________GCCUGCAAUGAAAACAAAAAGCGAUUUGCAAGUUUUCUAUUGCGCAUAAAAGUAUGCUAUGGAAAC_AUUUUUUCCC_UG_CU .......((.......................(((((((((((.....((.....))..)))))).)))))(((((....)))))...))..................... (-15.03 = -15.03 + -0.00)


| Location | 16,941,892 – 16,941,986 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
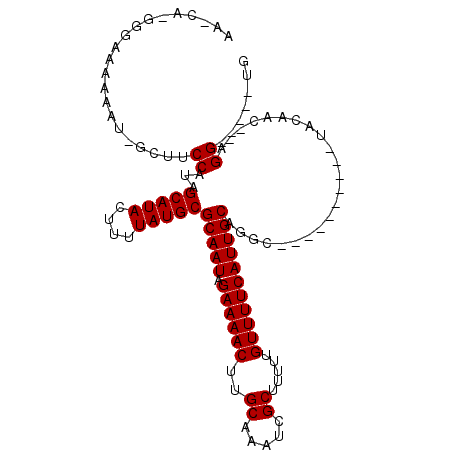
>2L_DroMel_CAF1 16941892 94 - 22407834 AAGCGGAGGGAAAAAUAGGUUCCAUAGCAUACUUUUAUGCGCAAUAGAAAACUUGCAAAUCGCUUUUUGUUUUCAUUGCAGGC----------UAAAAC--AGG-----CG .(((....((((.......))))...(((((....)))))(((((.((((((..((.....)).....)))))))))))..))----------).....--...-----.. ( -21.90) >DroVir_CAF1 69649 94 - 1 CGUCAUGGGAAAAAAU--UUUCCACAGCAUACUUUUAUGCGCAAUAGAAAACUUGCAAAUCGCUUUUUGUUUUCAUUGCAGGC----------AAC-----AGGACGAAUG ((((.(((((((....--))))).))(((((....)))))(((((.((((((..((.....)).....))))))))))).(..----------..)-----..)))).... ( -26.50) >DroPse_CAF1 56412 98 - 1 AA----GUGAAAAAAUGGCUUCCAUAGCAUACUUUUAUGCGCAAUAGAAAACUUGCAAAUCGCUUUUUGUUUUCAUUGCAGCC--AGGAUCCAUCCAAU--GGG-----UG ..----.........(((((......(((((....)))))(((((.((((((..((.....)).....)))))))))))))))--)..((((((...))--)))-----). ( -27.10) >DroGri_CAF1 58357 99 - 1 CGUCAUGGGAAAAAAU--UUCCCACAGCAUACUUUUAUGCGCAAUAGAAAACUUGCAAAUCGCUUUUUGUUUUCAUUGCCGGC----------AAGGAGUGAGGAGGGGUG .((..((((((.....--))))))..)).(((((((.((((((((.((((((..((.....)).....)))))))))))..))----------)))))))).......... ( -27.30) >DroAna_CAF1 50652 99 - 1 UGGCAGGGG-AAAAAUAGGAUCCAUAGCAUACUUUUAUGCGCAAUAGAAAACUUGCAAAUCGCUUUUUGUUUUCAUUGCAGGCCGAGGA----UAAAAC--AGG-----CA .(((...((-(.........)))...(((((....)))))(((((.((((((..((.....)).....)))))))))))..))).....----......--...-----.. ( -22.90) >DroPer_CAF1 57046 98 - 1 AA----GUGAAAAAAUGGCUUCCAUAGCAUACUUUUAUGCGCAAUAGAAAACUUGCAAAUCGCUUUUUGUUUUCAUUGCAGCC--AGGAUCCAUCCAAU--GGG-----UG ..----.........(((((......(((((....)))))(((((.((((((..((.....)).....)))))))))))))))--)..((((((...))--)))-----). ( -27.10) >consensus AA_CA_GGGAAAAAAU_GCUUCCAUAGCAUACUUUUAUGCGCAAUAGAAAACUUGCAAAUCGCUUUUUGUUUUCAUUGCAGGC__________UACAAC__AGG_____UG .....................((...(((((....)))))(((((.((((((..((.....)).....))))))))))).......................))....... (-17.38 = -17.38 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:54 2006