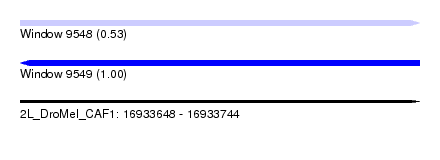
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,933,648 – 16,933,744 |
| Length | 96 |
| Max. P | 0.999433 |

| Location | 16,933,648 – 16,933,744 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -19.87 |
| Energy contribution | -19.82 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
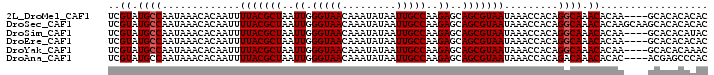
>2L_DroMel_CAF1 16933648 96 + 22407834 GUGUGUGUGC----UUGUGUUUGCCUGUGGUUUAUUACGCUGCUCUUGGCAAUUAUAUUUGUUACCCAAUUAGCGUAAAAUUGUGUUUAUUGGCAUACGA ...(((((((----(.(((...((..(..((((..(((((((....((((((......))))))......)))))))))))..))).))).)))))))). ( -23.40) >DroSec_CAF1 46250 100 + 1 GUGUGUGUGCUUGCUUGUGUUUGCCUGUGGUUUAUUACGCUGCUCUUGGCAAUUAUAUUUGUUACCCAAUUAGCGUAAAAUUGUGUUUAUUGGCAUACGA ...(((((((.....((((.(((((.(..((.......))..)....))))).)))).........((((.((((........)))).))))))))))). ( -22.70) >DroSim_CAF1 51627 96 + 1 GUAUGUGUGC----UUGUGUUUGCCUGUGGUUUAUUACGCUGCUCUUGGCAAUUAUAUUUGUUACCCAAUUAGCGUAAAAUUGUGUUUAUUGGCAUACGA ...(((((((----(.(((...((..(..((((..(((((((....((((((......))))))......)))))))))))..))).))).)))))))). ( -23.50) >DroEre_CAF1 49007 96 + 1 GUGUGUGUGC----UUGUGUUUGCCUGUGGUUUAUUACGCUGCUCUUGGCAAUUAUAUUUGUUACCCAAUUAGCGUAAAAUUGUGUUUAUUGGCAUACGA ...(((((((----(.(((...((..(..((((..(((((((....((((((......))))))......)))))))))))..))).))).)))))))). ( -23.40) >DroYak_CAF1 46245 96 + 1 GUUUGUGUGC----UUGUGUUUGCCUGUGGUUUAUUACGCUGCUCUUGGCAAUUAUAUUUGUUACCCAAUUAGCGUAAAAUUGUGUUUAUUGGCAUACGA ..((((((((----(.(((...((..(..((((..(((((((....((((((......))))))......)))))))))))..))).))).))))))))) ( -23.90) >DroAna_CAF1 43718 96 + 1 GUGGGCUCGU----GUGUGUUUGUCUGUGGUUUAUUACGCUGCUCUUGGCAAUUAUAUUUGUUACCCAAUUAGCGUAAAAUUGUGUUUAUUGGCAUACGA ......((((----((((...((..((..((((..(((((((....((((((......))))))......)))))))))))..))..))...)))))))) ( -22.30) >consensus GUGUGUGUGC____UUGUGUUUGCCUGUGGUUUAUUACGCUGCUCUUGGCAAUUAUAUUUGUUACCCAAUUAGCGUAAAAUUGUGUUUAUUGGCAUACGA ((((((.........((((.(((((.(..((.......))..)....))))).)))).........((((.((((........)))).)))))))))).. (-19.87 = -19.82 + -0.05)

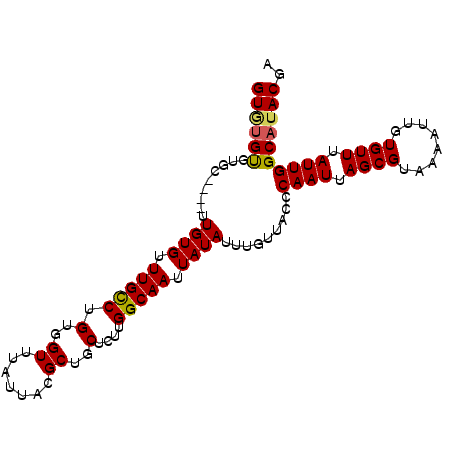
| Location | 16,933,648 – 16,933,744 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -16.24 |
| Consensus MFE | -15.67 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16933648 96 - 22407834 UCGUAUGCCAAUAAACACAAUUUUACGCUAAUUGGGUAACAAAUAUAAUUGCCAAGAGCAGCGUAAUAAACCACAGGCAAACACAA----GCACACACAC ..((.((((.............(((((((..((.(((((.........)))))..))..))))))).........)))).))....----.......... ( -16.95) >DroSec_CAF1 46250 100 - 1 UCGUAUGCCAAUAAACACAAUUUUACGCUAAUUGGGUAACAAAUAUAAUUGCCAAGAGCAGCGUAAUAAACCACAGGCAAACACAAGCAAGCACACACAC ..((.((((.............(((((((..((.(((((.........)))))..))..))))))).........)))).)).................. ( -16.95) >DroSim_CAF1 51627 96 - 1 UCGUAUGCCAAUAAACACAAUUUUACGCUAAUUGGGUAACAAAUAUAAUUGCCAAGAGCAGCGUAAUAAACCACAGGCAAACACAA----GCACACAUAC ..((.((((.............(((((((..((.(((((.........)))))..))..))))))).........)))).))....----.......... ( -16.95) >DroEre_CAF1 49007 96 - 1 UCGUAUGCCAAUAAACACAAUUUUACGCUAAUUGGGUAACAAAUAUAAUUGCCAAGAGCAGCGUAAUAAACCACAGGCAAACACAA----GCACACACAC ..((.((((.............(((((((..((.(((((.........)))))..))..))))))).........)))).))....----.......... ( -16.95) >DroYak_CAF1 46245 96 - 1 UCGUAUGCCAAUAAACACAAUUUUACGCUAAUUGGGUAACAAAUAUAAUUGCCAAGAGCAGCGUAAUAAACCACAGGCAAACACAA----GCACACAAAC ..((.((((.............(((((((..((.(((((.........)))))..))..))))))).........)))).))....----.......... ( -16.95) >DroAna_CAF1 43718 96 - 1 UCGUAUGCCAAUAAACACAAUUUUACGCUAAUUGGGUAACAAAUAUAAUUGCCAAGAGCAGCGUAAUAAACCACAGACAAACACAC----ACGAGCCCAC ((((.((...............(((((((..((.(((((.........)))))..))..))))))).........(.....).)).----))))...... ( -12.70) >consensus UCGUAUGCCAAUAAACACAAUUUUACGCUAAUUGGGUAACAAAUAUAAUUGCCAAGAGCAGCGUAAUAAACCACAGGCAAACACAA____GCACACACAC ..((.((((.............(((((((..((.(((((.........)))))..))..))))))).........)))).)).................. (-15.67 = -15.83 + 0.17)
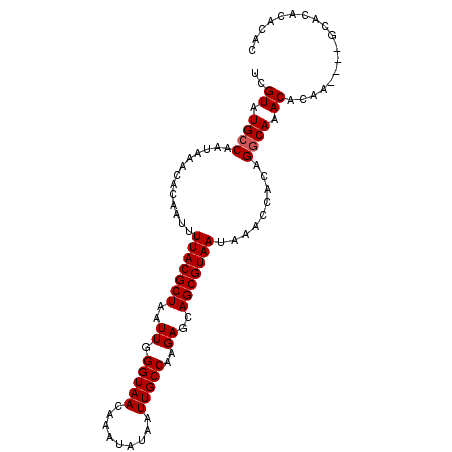
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:50 2006