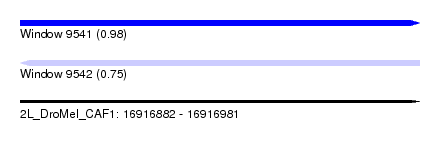
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,916,882 – 16,916,981 |
| Length | 99 |
| Max. P | 0.984348 |

| Location | 16,916,882 – 16,916,981 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -18.14 |
| Energy contribution | -19.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16916882 99 + 22407834 AAUUUCAUAAUUAUGCAAGCACAAAUGCCAUUGUGUGACCUCCCCACCCCACUUGUGGGGGCAUAGUCCACACAAGCCCCACCCCCA---CCACCUGCUUAA .........(((((((..((((((......)))))).....((((((.......)))))))))))))......((((..........---......)))).. ( -23.29) >DroSec_CAF1 29734 98 + 1 AAUUUCAUAAUUAUGCAAGCACAAAUGCCAUUGUGUGACCUCCCCAC-CCACUUGUGGGGGCAUAGUCCACACAAGUCCCUCUCCCA---CCGCCUGCUUAA ..............(((.((..........(((((((((.(((((((-......)))))))....)).)))))))(.....).....---..)).))).... ( -23.80) >DroSim_CAF1 34398 98 + 1 AAUUUCAUAAUUAUGCAAGCACAAAUGCCAUUGUGUGACCUCCCCAC-CCACUUGUGGGGGCAUGGUCCACACAAGUCCCUCCCCCA---CCGCCUGCUUAA ..............(((.((.(....)...(((((((((((((((((-......)))))))...))).)))))))............---..)).))).... ( -27.20) >DroYak_CAF1 29237 98 + 1 AAUUUCAUAAUUAUGCAAGCACAAAUGCCAUUGUGUGACCUCCCCAC-CCACUUGUGUG-CAUCGGUCCGC-CACGCACCAUC-UCAUCCCCCUUUGCUUAA ..............((((((((((......))))))...........-.....((.(((-(...((....)-)..))))))..-..........)))).... ( -15.90) >consensus AAUUUCAUAAUUAUGCAAGCACAAAUGCCAUUGUGUGACCUCCCCAC_CCACUUGUGGGGGCAUAGUCCACACAAGCCCCACCCCCA___CCGCCUGCUUAA ..............(((.(((....)))..(((((((((((((((((.......)))))))...))).)))))))....................))).... (-18.14 = -19.70 + 1.56)

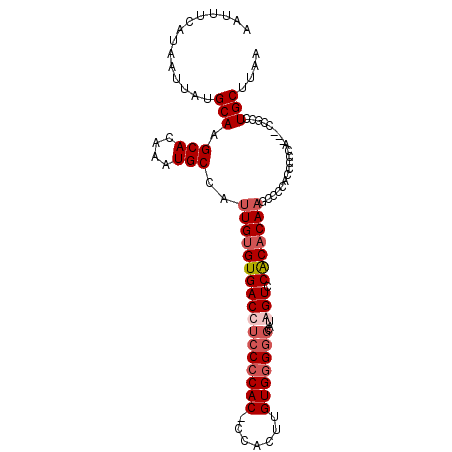
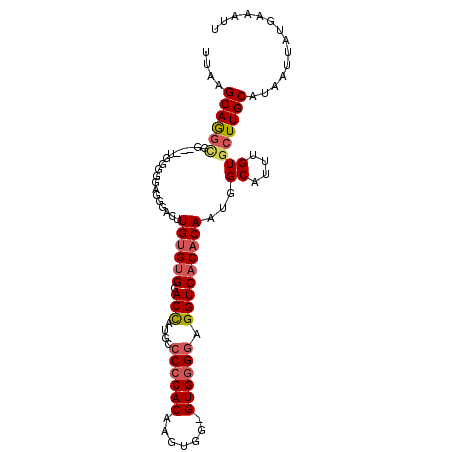
| Location | 16,916,882 – 16,916,981 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -22.36 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16916882 99 - 22407834 UUAAGCAGGUGG---UGGGGGUGGGGCUUGUGUGGACUAUGCCCCCACAAGUGGGGUGGGGAGGUCACACAAUGGCAUUUGUGCUUGCAUAAUUAUGAAAUU ...........(---..((.(..(((((((((((..((.(.((((((....))))).).).))..))))))..))).))..).))..).............. ( -32.70) >DroSec_CAF1 29734 98 - 1 UUAAGCAGGCGG---UGGGAGAGGGACUUGUGUGGACUAUGCCCCCACAAGUGG-GUGGGGAGGUCACACAAUGGCAUUUGUGCUUGCAUAAUUAUGAAAUU ....((((((..---............(((((((..((....((((((......-))))))))..)))))))..((....)))))))).............. ( -31.40) >DroSim_CAF1 34398 98 - 1 UUAAGCAGGCGG---UGGGGGAGGGACUUGUGUGGACCAUGCCCCCACAAGUGG-GUGGGGAGGUCACACAAUGGCAUUUGUGCUUGCAUAAUUAUGAAAUU ....((((((.(---((((((.((..(......)..))...)))))))..((((-.(.....).))))((((......)))))))))).............. ( -35.00) >DroYak_CAF1 29237 98 - 1 UUAAGCAAAGGGGGAUGA-GAUGGUGCGUG-GCGGACCGAUG-CACACAAGUGG-GUGGGGAGGUCACACAAUGGCAUUUGUGCUUGCAUAAUUAUGAAAUU ....((((..(.(((((.-.....((.(((-((...((.((.-(((....))).-)).))...))))).))....))))).)..)))).............. ( -26.50) >consensus UUAAGCAGGCGG___UGGGGGAGGGACUUGUGUGGACCAUGCCCCCACAAGUGG_GUGGGGAGGUCACACAAUGGCAUUUGUGCUUGCAUAAUUAUGAAAUU ....((((((..................((((((.(((....((((((.......)))))).)))))))))...((....)))))))).............. (-22.36 = -23.30 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:44 2006