
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,899,937 – 16,900,078 |
| Length | 141 |
| Max. P | 0.950991 |

| Location | 16,899,937 – 16,900,057 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -21.10 |
| Energy contribution | -20.97 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16899937 120 - 22407834 CUUGAUCUUGGGAUUUCUCUGAAUCUUCUCGUUAUCCAUGGCCUCAGCCAUAAGACAAGCAGCCACUGCCGUUUGUCUCGUGUGCAGAGCCUUCAUCAAUGAUACUAUUUUCAUGGCCAG ...(((..(((((.(((...)))...)))))..)))..(((((...((((..((((((((.((....)).))))))))..)).)).............((((........))))))))). ( -34.10) >DroVir_CAF1 11682 120 - 1 CUUGAUCUUUGGAUUGCGCUGGAUCUUCUCGUUUUCCAUGGCGUCCGCCAUGAGGCAGGCGGCGACAGCUGUUUGGCGCGUGUGCAGCACCUUCAUAAGGGACACAAUUUUGAUGGCCAG .........((((..(((..((.....)))))..))))((((((((((((((.(.((((((((....)))))))).).)))).)).....((.....)))))).((....))...)))). ( -39.00) >DroGri_CAF1 11131 120 - 1 CUUUAUCUUUGGAUUGCGCUGAAUCUUCUCGUUUUCCAUGGCAUCGGCCAUGAGGCAGGCAGCAACAGCUGUUUGCCGUGUGUGCAACACUUUGAGCAGCGAGACUAUUUUAAUGGCCAG .........(((........((((..((((((((((((((((....)))))).((((((((((....))))))))))((((.....))))...))).)))))))..))))......))). ( -43.74) >DroWil_CAF1 10280 120 - 1 CUUGAUCUUGGGAUUUCUCUGAAUCUUCUCAUUAUCCAUGGCCUCGGCCAUGAGGCAAGCCGCCACAGCCGUUUGUCGUGUAUGCAAAGCCUUGCAGAGUGACACGAUCUUGAUGGCCAA ...(((..(((((.(((...)))...)))))..)))((((((....)))))).(((.....)))...((((((.(((((((.(((((....))))).....)))))))...))))))... ( -42.40) >DroMoj_CAF1 10542 120 - 1 CUUUAUCUUUGGGUUUCGCUGAAUUUUCUCAUUCUCCAUGGCAUCUGCCAUGAGGCAGGCAGCAACAGCCGUCUGUCUUGUAUGCAGCACCUUCAUCAGGGAAACAAUUUUCAUGGCUAG ............(((((((((...............((((((....))))))((((((((.((....)).))))))))......)))).(((.....))))))))............... ( -36.00) >DroAna_CAF1 10355 120 - 1 CUUAAUUUUGGGAUUCCUCUGGAUCUUCUCAUUGUCCAUUGCCUCCGCCAUGAGGCAGGCAGCCACCGCCGUUUGUCUCGUGUGGAGGGCUUUCAUGAGAGCCACUAUUUUGAUGGCCAG ........(((((.(((...)))...))))).((.(((((..((((((.(((((((((((.((....)).)))))))))))))))))((((((....))))))........))))).)). ( -46.40) >consensus CUUGAUCUUGGGAUUUCGCUGAAUCUUCUCAUUAUCCAUGGCCUCCGCCAUGAGGCAGGCAGCCACAGCCGUUUGUCGCGUGUGCAGCACCUUCAUCAGGGACACUAUUUUGAUGGCCAG .................((((...............((((((....)))))).(((((((.((....)).))))))).......))))................................ (-21.10 = -20.97 + -0.14)
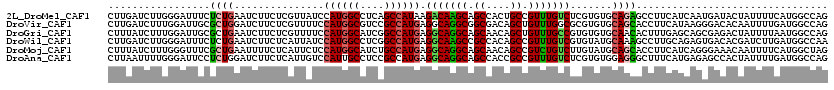
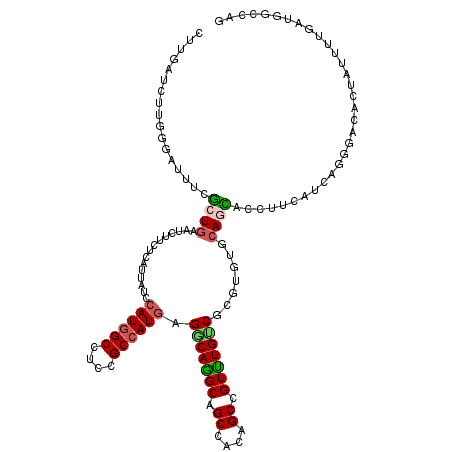
| Location | 16,899,977 – 16,900,078 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16899977 101 + 22407834 CGAGACAAACGGCAGUGGCUGCUUGUCUUAUGGCUGAGGCCAUGGAUAACGAGAAGAUUCAGAGAAAUCCCAAGAUCAAGAAGGUA----ACAAAAGAGGUAUAA .(((((((.((((....)))).)))))))(((((....)))))............((((..((....))....)))).........----............... ( -22.50) >DroPse_CAF1 11891 104 + 1 CGCGGCAGACGGCGGUGGCCGCCUGUCUCAUGGCGGAGGCCUUGGACAAUGAGAAGAUCCAACGGAAUCCCAAGAUCAAGAAGGUGAGGU-CGACAAACGUAUAC .(.(((((.((((....)))).))))).).(((((((..((((((((........).))))).))..))).....(((......))).))-))............ ( -29.90) >DroGri_CAF1 11171 104 + 1 CACGGCAAACAGCUGUUGCUGCCUGCCUCAUGGCCGAUGCCAUGGAAAACGAGAAGAUUCAGCGCAAUCCAAAGAUAAAGAAGGUAGGUA-GGAAUGGAGAGUAA ........((..(((((.((((((((((((((((....))))))...........((((......))))............)))))))))-).)))))...)).. ( -30.50) >DroWil_CAF1 10320 104 + 1 CACGACAAACGGCUGUGGCGGCUUGCCUCAUGGCCGAGGCCAUGGAUAAUGAGAAGAUUCAGAGAAAUCCCAAGAUCAAGAAGGUGAGUU-UGUUAAUUGUGUUC (((((..((((((....))((((..(((((((((....))))))...........((((..((....))....))))....)))..))))-))))..)))))... ( -30.00) >DroMoj_CAF1 10582 101 + 1 CAAGACAGACGGCUGUUGCUGCCUGCCUCAUGGCAGAUGCCAUGGAGAAUGAGAAAAUUCAGCGAAACCCAAAGAUAAAGAAGGUGGGUA---AAUGCAGCGUA- ...........(((((...((((..(((((((((....))))))..((((......)))).....................)))..))))---...)))))...- ( -32.90) >DroPer_CAF1 11909 104 + 1 CGCGGCAGACGGCGGUGGCCGCCUGUCUCAUGGCGGAGGCCUUGGACAAUGAGAAGAUCCAACGGAAUCCCAAGAUCAAGAAGGUGAGGU-CGACAAACGAAUAC .(.(((((.((((....)))).))))).).(((((((..((((((((........).))))).))..))).....(((......))).))-))............ ( -29.90) >consensus CACGACAAACGGCUGUGGCUGCCUGCCUCAUGGCCGAGGCCAUGGACAAUGAGAAGAUUCAGCGAAAUCCCAAGAUCAAGAAGGUGAGUU_CGAAAAAAGUAUAC ...(((((.((((....)))).)))))(((((((....)))))))............................................................ (-19.23 = -19.32 + 0.09)

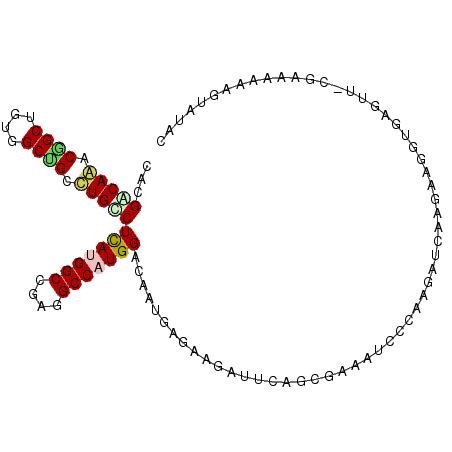

| Location | 16,899,977 – 16,900,078 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -28.69 |
| Consensus MFE | -20.24 |
| Energy contribution | -19.93 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16899977 101 - 22407834 UUAUACCUCUUUUGU----UACCUUCUUGAUCUUGGGAUUUCUCUGAAUCUUCUCGUUAUCCAUGGCCUCAGCCAUAAGACAAGCAGCCACUGCCGUUUGUCUCG ...............----.........(((..(((((.(((...)))...)))))..))).(((((....))))).((((((((.((....)).)))))))).. ( -21.90) >DroPse_CAF1 11891 104 - 1 GUAUACGUUUGUCG-ACCUCACCUUCUUGAUCUUGGGAUUCCGUUGGAUCUUCUCAUUGUCCAAGGCCUCCGCCAUGAGACAGGCGGCCACCGCCGUCUGCCGCG .....(((......-..((((.......(((..(((((.(((...)))...)))))..)))...(((....))).)))).((((((((....))))))))..))) ( -30.40) >DroGri_CAF1 11171 104 - 1 UUACUCUCCAUUCC-UACCUACCUUCUUUAUCUUUGGAUUGCGCUGAAUCUUCUCGUUUUCCAUGGCAUCGGCCAUGAGGCAGGCAGCAACAGCUGUUUGCCGUG ..............-....................(((..(((..((....)).)))..)))(((((....)))))..((((((((((....))))))))))... ( -31.10) >DroWil_CAF1 10320 104 - 1 GAACACAAUUAACA-AACUCACCUUCUUGAUCUUGGGAUUUCUCUGAAUCUUCUCAUUAUCCAUGGCCUCGGCCAUGAGGCAAGCCGCCACAGCCGUUUGUCGUG ...(((........-.............(((..(((((.(((...)))...)))))..)))((((((....)))))).(((((((.((....)).)))))))))) ( -28.80) >DroMoj_CAF1 10582 101 - 1 -UACGCUGCAUU---UACCCACCUUCUUUAUCUUUGGGUUUCGCUGAAUUUUCUCAUUCUCCAUGGCAUCUGCCAUGAGGCAGGCAGCAACAGCCGUCUGUCUUG -......((...---.(((((.............)))))...)).((((......))))..((((((....))))))((((((((.((....)).)))))))).. ( -29.32) >DroPer_CAF1 11909 104 - 1 GUAUUCGUUUGUCG-ACCUCACCUUCUUGAUCUUGGGAUUCCGUUGGAUCUUCUCAUUGUCCAAGGCCUCCGCCAUGAGACAGGCGGCCACCGCCGUCUGCCGCG ....(((.....))-).((((.......(((..(((((.(((...)))...)))))..)))...(((....))).)))).((((((((....))))))))..... ( -30.60) >consensus GUACACGUCUUUCG_AACUCACCUUCUUGAUCUUGGGAUUUCGCUGAAUCUUCUCAUUAUCCAUGGCCUCCGCCAUGAGACAGGCAGCCACAGCCGUCUGCCGCG ...................................((((((....))))))..........((((((....)))))).(((((((.((....)).)))))))... (-20.24 = -19.93 + -0.30)

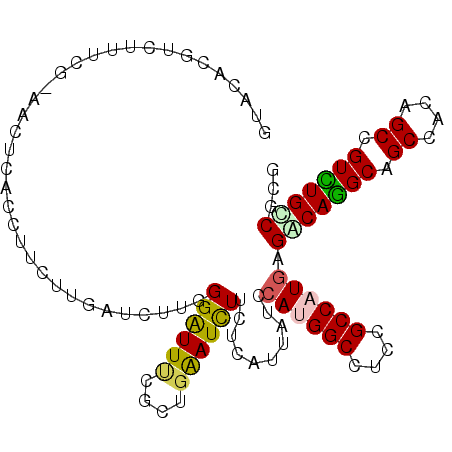
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:35 2006