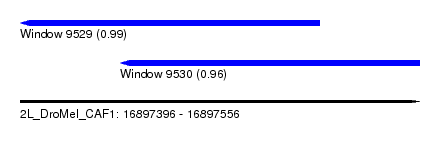
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,897,396 – 16,897,556 |
| Length | 160 |
| Max. P | 0.990984 |

| Location | 16,897,396 – 16,897,516 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -39.02 |
| Energy contribution | -40.70 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
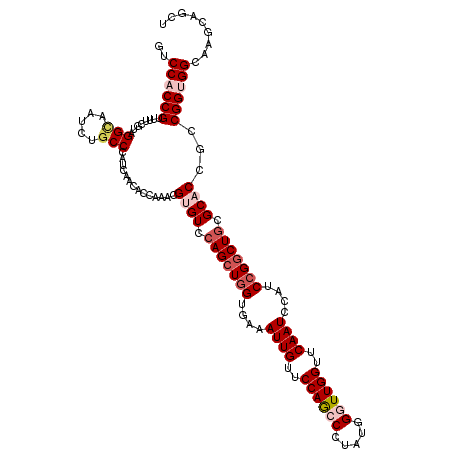
>2L_DroMel_CAF1 16897396 120 - 22407834 CGUGUACAGCUGGUGAAAUUGUUCCAGCCAUAUGGGUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGUGGUAAGCAGCUUUUCAAGCACAAACAGCGCAAGGUAGCCGGCUCCCUGAAC .(((..(((((((....((((..((((((.....))))))..))))....)))))))..))).((((((........(((.....)))........(....)...))))))......... ( -47.10) >DroSec_CAF1 10104 120 - 1 CGUGUCCAGCUGGUGAAAUUGUUCCAGCCCUAUGGAUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGGGGCAAGCAGCUUUUCAAGCACAAACAGCGCAAGGUAGCCGGCUCCCUGAAC .((((.(((((((....((((..((((((....)).))))..))))....))))))).))))....((((((..((.(((((....((.......))..))))).))..))))))..... ( -46.40) >DroSim_CAF1 12375 120 - 1 CGUGUCCAGCUGGUGAAAUUGUUCCAGCCCUAUGGAUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGGGGCAAGCAGCUUUUCAAGCACAAACAGCGCAAGGUAGCCGGCUCCCUGAAC .((((.(((((((....((((..((((((....)).))))..))))....))))))).))))....((((((..((.(((((....((.......))..))))).))..))))))..... ( -46.40) >DroEre_CAF1 12028 120 - 1 CGCGUCCAGCUAGUGAAAUUGUUCCAGCCCUACGGGUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGUGGCAAGCAGCUUUUUAAGCACAAACAGCGCAAGGGAGCCGGCUCCCUAAAC .((((...(((......((((..(((((((...)))))))..))))(((.(((((........))))))))..))).(((.....))).......)))).((((((....)))))).... ( -47.90) >DroYak_CAF1 9942 120 - 1 CGCGUCCAGCUGGUGAAAUUGUUCCAACCCUAUGGGUUGGUUCAAUCCAUCCGUCUUCGCACCGCCGGUGGCAAGCAGCUUUUCAAGCACAAACAGCGCAAGGUAGCCGGCUCCCUAAAC .(((....((.((((..((((..(((((((...)))))))..)))).)))).))...)))...((((((.((.....(((.....)))........(....))).))))))......... ( -40.60) >consensus CGUGUCCAGCUGGUGAAAUUGUUCCAGCCCUAUGGGUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGUGGCAAGCAGCUUUUCAAGCACAAACAGCGCAAGGUAGCCGGCUCCCUGAAC .((((.(((((((....((((..((((((.....))))))..))))....))))))).)))).((((((.((.....(((.....)))........(....))).))))))......... (-39.02 = -40.70 + 1.68)


| Location | 16,897,436 – 16,897,556 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -34.50 |
| Energy contribution | -36.18 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16897436 120 - 22407834 GUCCACCGUUUUCGUGGGGAAUUUGCCAAUCAACACCAAACGUGUACAGCUGGUGAAAUUGUUCCAGCCAUAUGGGUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGUGGUAAGCAGCU ..(((((((((..(((..((.........))..))).))))(((..(((((((....((((..((((((.....))))))..))))....)))))))..)))....)))))......... ( -46.10) >DroSec_CAF1 10144 120 - 1 GUCCACCGUUUUCGUAGGUAAUCUGCCCAUCAACACCAAACGUGUCCAGCUGGUGAAAUUGUUCCAGCCCUAUGGAUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGGGGCAAGCAGCU ((((.(((.....((((.....))))...............((((.(((((((....((((..((((((....)).))))..))))....))))))).))))...)))))))........ ( -39.50) >DroSim_CAF1 12415 120 - 1 GUCCACCGUUUUCGUAGGUAAUCUGCCCAUCAACACCAAACGUGUCCAGCUGGUGAAAUUGUUCCAGCCCUAUGGAUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGGGGCAAGCAGCU ((((.(((.....((((.....))))...............((((.(((((((....((((..((((((....)).))))..))))....))))))).))))...)))))))........ ( -39.50) >DroEre_CAF1 12068 120 - 1 GUCCACCGUUUUCGUAGGCAAUCUGCCCAUUAACACCAAACGCGUCCAGCUAGUGAAAUUGUUCCAGCCCUACGGGUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGUGGCAAGCAGCU (.((((((........(((.....)))..............(((..(((((.(((..((((..(((((((...)))))))..)))).)))..)))))))).....)))))))........ ( -40.70) >DroYak_CAF1 9982 120 - 1 AUCCACCGUUUUCGUGGGCAAUCUGCCCAUAAACACCAAACGCGUCCAGCUGGUGAAAUUGUUCCAACCCUAUGGGUUGGUUCAAUCCAUCCGUCUUCGCACCGCCGGUGGCAAGCAGCU ..(((((((((..((((((.....)))))))))).......(((....((.((((..((((..(((((((...)))))))..)))).)))).))...)))......)))))......... ( -43.80) >consensus GUCCACCGUUUUCGUAGGCAAUCUGCCCAUCAACACCAAACGUGUCCAGCUGGUGAAAUUGUUCCAGCCCUAUGGGUUGGUUCAAUCCAUCCGGCUGCGCACCGCCGGUGGCAAGCAGCU ..((((((........(((.....)))..............((((.(((((((....((((..((((((.....))))))..))))....))))))).))))...))))))......... (-34.50 = -36.18 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:32 2006