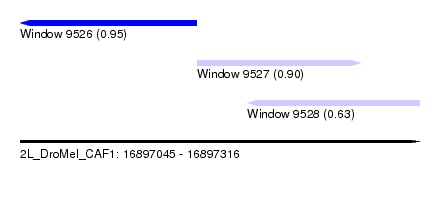
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,897,045 – 16,897,316 |
| Length | 271 |
| Max. P | 0.948839 |

| Location | 16,897,045 – 16,897,165 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -44.44 |
| Consensus MFE | -37.96 |
| Energy contribution | -38.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
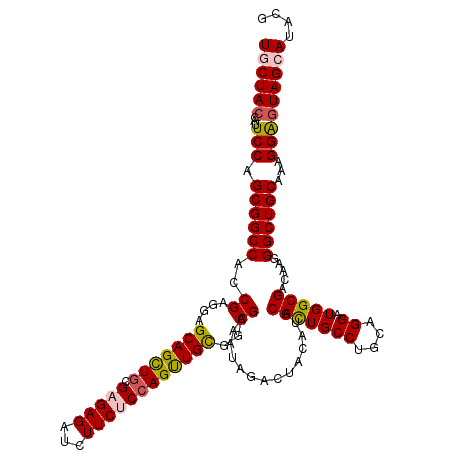
>2L_DroMel_CAF1 16897045 120 - 22407834 UGCUACACAUUCCAGCGGCCACCGAGGAGCAGCUGCGAGAGAUCUUCUCCAGUUGCGGCGAAAUAGACUACAUUCGCUGCCUGCAGGAUGGCGACAAGGGCUGCAAAGGAGUAGCAUACG ((((((....(((.((((((.((.(((.(((((((.(((((...))))))))))))((((((..........)))))).)))...)).((....))..))))))...))))))))).... ( -47.50) >DroSec_CAF1 9753 120 - 1 UGCUACUCAUUCCAGCGGCCACCGAGGAGCAGCUGCGAGAGAUCUUCUCAAGUUGUGGCGAAAUAGACUACAUACGCUGCCUGCAGGAUGGCGACAAGGGCUGCAAAGGAGUAGCAUACG ((((((((......((((((.((..((.(((((...(((((...)))))..((((((((......).))))).)))))))))...)).((....))..))))))....)))))))).... ( -46.90) >DroSim_CAF1 12024 120 - 1 UGCUACUCAUUCCAGCGGCCACCGAGGAGCAGCUGCGAGAGAUCUUCUCCAGUUGUGGCGAAAUAGACUACAUACGUUGCCUGCAGGAUGGCGACAAGGGCUGCAAAGGAGUAGCAUACG ((((((((......((((((..((....(((((((.(((((...))))))))))))..))...............((((((........))))))...))))))....)))))))).... ( -49.00) >DroEre_CAF1 11680 117 - 1 U-CUAC--CAUCCAGCGGCCAACGAGGAGCAGUUGCGCGAGAUCUUCUCCAGCUGCGGCGAGAUAGACUACAUACGCUGCCUGCAGGAUGGCGACAAGGGCUGCAAAGGGGUAGCAUACG .-((((--(.....((((((..((....(((((((...(((.....))))))))))..))..............(((((((....)).))))).....)))))).....)))))...... ( -39.80) >DroYak_CAF1 9585 117 - 1 U-CUAA--UAUCCAGCGGCCACCGAGGAGCAGCUGCGCGAGAUCUUCUCCAGCUGCGGCGAGAUAGACUACAUACGCUGCCUGCAGGAUGGCGACAAGGGCUGCAAAGGAGUAGCAUACG .-(((.--..(((.((((((..((....(((((((...(((.....))))))))))..))..............(((((((....)).))))).....))))))...))).)))...... ( -39.00) >consensus UGCUAC_CAUUCCAGCGGCCACCGAGGAGCAGCUGCGAGAGAUCUUCUCCAGUUGCGGCGAAAUAGACUACAUACGCUGCCUGCAGGAUGGCGACAAGGGCUGCAAAGGAGUAGCAUACG ((((((....(((.((((((..((....(((((((.(((((...))))))))))))..))..............(((((((....)).))))).....))))))...))))))))).... (-37.96 = -38.20 + 0.24)

| Location | 16,897,165 – 16,897,276 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.895071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16897165 111 + 22407834 CAUGAUGUUAGCAUUUAGUUAAAACUACUUACU--G-------CUUUACUUACAGUACUUUAGGCUGCCAACGAAAAUGGUUCGCUUGGCAUCCUUGUCACUGGGCUGCUUAUCCUUUGC ...((((.((((...((((....))))......--.-------.........((((((...(((.((((((((((.....)))).)))))).))).)).)))).))))..))))...... ( -27.40) >DroSec_CAF1 9873 111 + 1 CUUAACGUUAGCACUUAGUUAAAACUACUGACU--U-------CUUUACUUACAGUACUUUAGGCUGCCAACGAAAAUGGUUCGCUUGGCAUCCUUGUCACUGGGCUGCUGAUCCUUUCC ......(((((((((((((..(((.(((((...--.-------.........))))).)))(((.((((((((((.....)))).)))))).)))....)))))).)))))))....... ( -30.12) >DroSim_CAF1 12144 111 + 1 CUUAAUGUUAGCACUUAGUUCAAACUACUGACU--U-------CUUUACUUACAGUACUUUAGGCUGCCAACGAAAAUGGUUCGCUUGGCAUCCUUGUCACUGGGCUGCUGAUCCUUUCC ......(((((((((((((.(((..(((((...--.-------.........))))).....((.((((((((((.....)))).)))))).)))))..)))))).)))))))....... ( -30.02) >DroEre_CAF1 11797 111 + 1 UUUAAUAUUAGCACUUUGUUAAAACUACUCUAU--U-------UAUUACUUACAGUAUUUUAGGCUGCCCACGAAAAUGGUUCGCUUGGCAUCCUUGUCAUUGGGCUGUUCAUCCUUUCC .......(((((.....))))).......(((.--.-------...(((.....)))...)))((.(((((((((.....))))..(((((....))))).))))).))........... ( -18.30) >DroYak_CAF1 9702 120 + 1 UUUAAUGUUAGCAUGUAGUUAAAACUACUCAAAAGUCAAGGAACGAUACUUACAGUAUUUUAGGCUGCCCACGAAUAUGGUUCGCUUGGCAUCCUUGUCACUGGGCUGCUGAUCCGCUCC .........(((..(((((....)))))...........(((.((((((.....)))))...(((.((((((((((...)))))..(((((....))))).))))).)))).)))))).. ( -30.80) >consensus CUUAAUGUUAGCACUUAGUUAAAACUACUCACU__U_______CUUUACUUACAGUACUUUAGGCUGCCAACGAAAAUGGUUCGCUUGGCAUCCUUGUCACUGGGCUGCUGAUCCUUUCC ......(((((((..((((....)))).........................((((((...(((.((((((((((.....)))).)))))).))).)).))))...)))))))....... (-23.34 = -23.62 + 0.28)

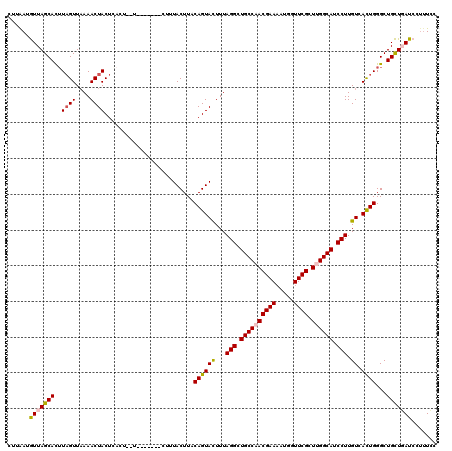
| Location | 16,897,199 – 16,897,316 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -32.20 |
| Energy contribution | -33.04 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
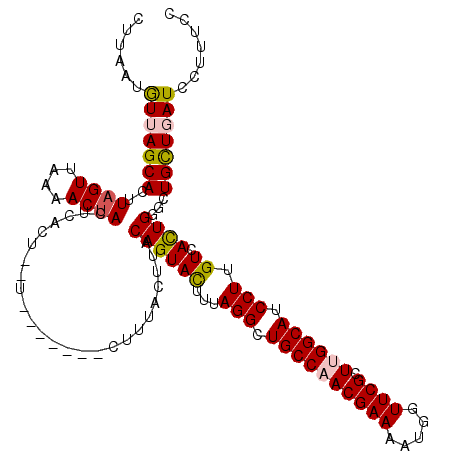
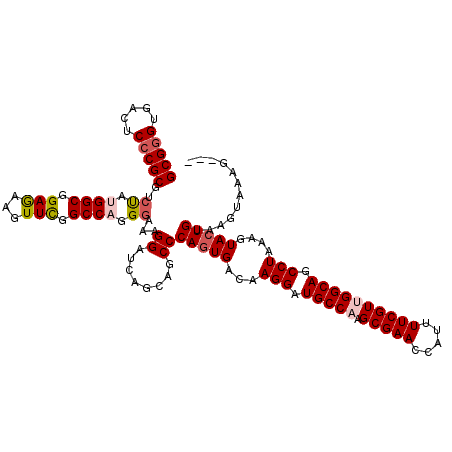
>2L_DroMel_CAF1 16897199 117 - 22407834 GCGGGUGACUCCCGCGUCUAUGGCGGAGAAAUUCGGCCAGGCAAAGGAUAAGCAGCCCAGUGACAAGGAUGCCAAGCGAACCAUUUUCGUUGGCAGCCUAAAGUACUGUAAGUAAAG--- (((((.....))))).....(((((((....))).))))(((............)))(((((...(((.((((((.((((.....)))))))))).)))....))))).........--- ( -40.90) >DroSec_CAF1 9907 117 - 1 GCGGGUGACGCCCGCGUCUAUGGCGGAGAAGUUCGGCCAGGGAAAGGAUCAGCAGCCCAGUGACAAGGAUGCCAAGCGAACCAUUUUCGUUGGCAGCCUAAAGUACUGUAAGUAAAG--- (((((.....))))).(((.((((.(((...))).)))).)))..............(((((...(((.((((((.((((.....)))))))))).)))....))))).........--- ( -41.10) >DroSim_CAF1 12178 117 - 1 GCGGGUGACUCCCGCGUCUAUGGCGGAGAAGUUCAGCCAGGGAAAGGAUCAGCAGCCCAGUGACAAGGAUGCCAAGCGAACCAUUUUCGUUGGCAGCCUAAAGUACUGUAAGUAAAG--- (((((.....))))).(((.((((.(((...))).)))).)))..............(((((...(((.((((((.((((.....)))))))))).)))....))))).........--- ( -38.60) >DroEre_CAF1 11831 117 - 1 GCGAGUUACUCCCGCGGCCAAGGCGGAAGGUUUUGGCCAAGGAAAGGAUGAACAGCCCAAUGACAAGGAUGCCAAGCGAACCAUUUUCGUGGGCAGCCUAAAAUACUGUAAGUAAUA--- ....((((((.((..(((((((((.....)))))))))..)).........((((..........(((.((((..(((((.....))))).)))).)))......)))).)))))).--- ( -34.99) >DroYak_CAF1 9742 120 - 1 GCGGGUUACUCCCGCGGCUAAGGCGGAGGGUUUUGGCCAGGGAGCGGAUCAGCAGCCCAGUGACAAGGAUGCCAAGCGAACCAUAUUCGUGGGCAGCCUAAAAUACUGUAAGUAUCGUUC ((.((((.(((((..(((((((((.....))))))))).)))))..)))).))....(((((...(((.((((..(((((.....))))).)))).)))....)))))............ ( -44.80) >consensus GCGGGUGACUCCCGCGUCUAUGGCGGAGAAGUUCGGCCAGGGAAAGGAUCAGCAGCCCAGUGACAAGGAUGCCAAGCGAACCAUUUUCGUUGGCAGCCUAAAGUACUGUAAGUAAAG___ (((((.....)))))..((.((((.(((...))).)))).))...((........))(((((...(((.(((((.(((((.....)))))))))).)))....)))))............ (-32.20 = -33.04 + 0.84)

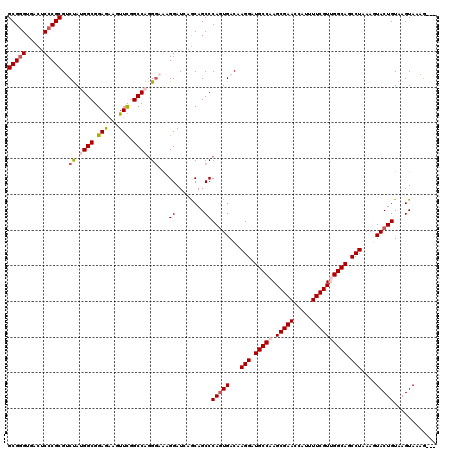
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:30 2006