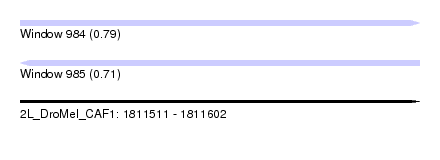
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,811,511 – 1,811,602 |
| Length | 91 |
| Max. P | 0.793517 |

| Location | 1,811,511 – 1,811,602 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1811511 91 + 22407834 UCCCCU--------UCCUCUGGAUCUCCUCCAAUAACUUGGGGAAUCUCCUAAUAUGGCAGAAGAACUGCCUCCGGUCGUGACAAUCUGUUCAGGUGGA ((((((--------.(..(((((..(((.((((....)))))))............(((((.....))))))))))..).(((.....))).))).))) ( -24.70) >DroSim_CAF1 8435 91 + 1 UCCACU--------UCCUCUGGAUCUCCUCCAAUAACUUGGGGGAUCUCCAUAUAUGGCAGAGGAACUGCCUCCGGUUGUGGCAAUAUGCUCAGGUGGC .(((((--------(((((((....((((((((....))))))))...(((....))))))))))..((((.........)))).........))))). ( -33.20) >DroEre_CAF1 19970 98 + 1 UCCACUUCCUCACUUCCUCUGGAUCUCCUCCAAUAACUUUGGGAGUCUCCA-AUAUGGCAGAGCAACUGCCUCCGGCCAGGACCAUCUGCUCAGGUGGA ..............(((((((((.((((..(((.....)))))))......-....(((((.....))))))))))..))))((((((....)))))). ( -28.90) >consensus UCCACU________UCCUCUGGAUCUCCUCCAAUAACUUGGGGAAUCUCCA_AUAUGGCAGAGGAACUGCCUCCGGUCGUGACAAUCUGCUCAGGUGGA .(((((........((..(((((..(((.((((....)))))))............(((((.....))))))))))....))...........))))). (-20.32 = -20.54 + 0.23)

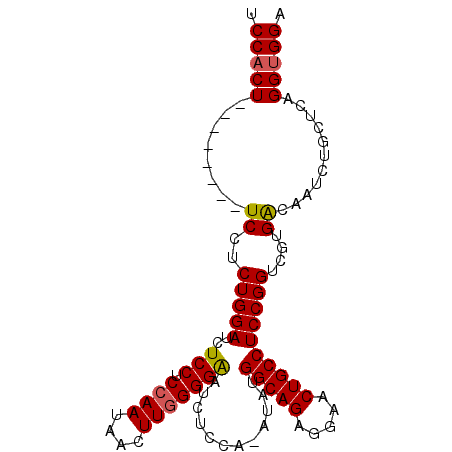
| Location | 1,811,511 – 1,811,602 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -23.80 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1811511 91 - 22407834 UCCACCUGAACAGAUUGUCACGACCGGAGGCAGUUCUUCUGCCAUAUUAGGAGAUUCCCCAAGUUAUUGGAGGAGAUCCAGAGGA--------AGGGGA (((.(((((........))....((...(((((.....)))))......(((..((((((((....)))).)))).)))...)).--------)))))) ( -28.40) >DroSim_CAF1 8435 91 - 1 GCCACCUGAGCAUAUUGCCACAACCGGAGGCAGUUCCUCUGCCAUAUAUGGAGAUCCCCCAAGUUAUUGGAGGAGAUCCAGAGGA--------AGUGGA .((((.........(((((.(.....).)))))(((((((((((....))).((((((((((....)))).)).)))))))))))--------))))). ( -34.80) >DroEre_CAF1 19970 98 - 1 UCCACCUGAGCAGAUGGUCCUGGCCGGAGGCAGUUGCUCUGCCAUAU-UGGAGACUCCCAAAGUUAUUGGAGGAGAUCCAGAGGAAGUGAGGAAGUGGA (((((((....))....((((.((....(((((.....)))))...(-((((..(((((((.....)))..)))).))))).....)).)))).))))) ( -31.20) >consensus UCCACCUGAGCAGAUUGUCACGACCGGAGGCAGUUCCUCUGCCAUAU_UGGAGAUUCCCCAAGUUAUUGGAGGAGAUCCAGAGGA________AGUGGA .((((....((.....)).....((...(((((.....)))))......(((..((((((((....)))).)))).)))...))..........)))). (-23.80 = -24.13 + 0.34)

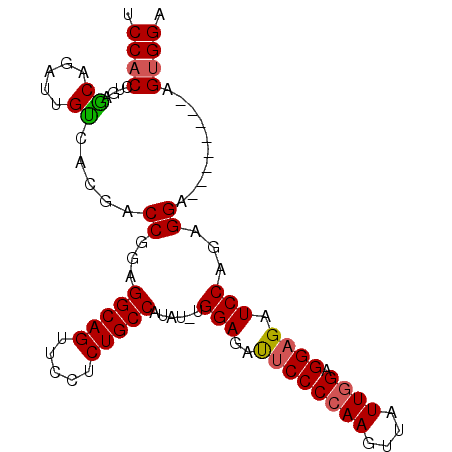
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:39 2006