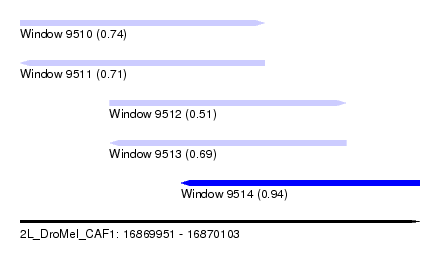
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,869,951 – 16,870,103 |
| Length | 152 |
| Max. P | 0.938842 |

| Location | 16,869,951 – 16,870,044 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.38 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16869951 93 + 22407834 CACAC---CACACAUGGGUAUAACGAAAUAAUGACGUUUGAUUGGCCUUCGGCAAGGCCGUUUG-------CUGUUUGUUGGCCAACUGUCAACGUCUUGUAA .((.(---((....)))))........((((.((((((.((((((((..(((((((....))))-------)))......)))))...))))))))))))).. ( -26.60) >DroSec_CAF1 9379 92 + 1 CACACCACCACACAUGGGUAAAGCGAAAUAAUGACGUUUGAUUGGCCUUCGGCAAGGCCGUUUG-------CUG----UUGGCCAACUGUCAACGUCUUGUAA .((.(((.......)))))........((((.((((((.((((((((..(((((((....))))-------)))----..)))))...))))))))))))).. ( -28.60) >DroSim_CAF1 11815 92 + 1 CACACCACCACACAUGGGUAAAGCGAAAUAAUGACGUUUGAUUGGCCUUCGGCAAGGCCGUUUG-------CUG----UUGGCCAACUGUCAACGUCUUGUAA .((.(((.......)))))........((((.((((((.((((((((..(((((((....))))-------)))----..)))))...))))))))))))).. ( -28.60) >DroEre_CAF1 41610 83 + 1 CA--------CACAUGGCCA-AACGAAAUAAUGACGUUUGAUUGGCCUUCGGCAAGGCCGUUUG-------CUG----UUGGCCAACUGUCAACGUCUUGUAA ..--------..........-......((((.((((((.((((((((..(((((((....))))-------)))----..)))))...))))))))))))).. ( -26.80) >DroYak_CAF1 4708 83 + 1 CA--------CACAUGGGAA-AACGAAAUAAUGACGUUUGAUUGGCCUUCGGCAAGGCCGUUUG-------CUG----UUGGCCAACUGUCAACGUCUUGUAA ..--------..........-......((((.((((((.((((((((..(((((((....))))-------)))----..)))))...))))))))))))).. ( -26.80) >DroPer_CAF1 34445 93 + 1 UAUACUACAAUAU------AUAACCAGAUAAUGACGUUUGAUUGGCCUUUGGCAAGGCCGUUUUAGCCUGUCGG----UUGGCCAAAUGUCAACGUCUUGUAA .............------...((.((((..(((((((((.(..(((...((((.(((.......)))))))))----)..).)))))))))..)))).)).. ( -28.70) >consensus CACAC___CACACAUGGGUA_AACGAAAUAAUGACGUUUGAUUGGCCUUCGGCAAGGCCGUUUG_______CUG____UUGGCCAACUGUCAACGUCUUGUAA ...........................((((.((((((.((((((((..((((...))))....................)))))...))))))))))))).. (-20.52 = -20.38 + -0.14)

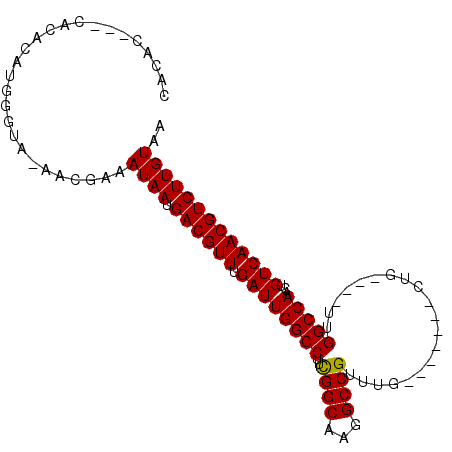

| Location | 16,869,951 – 16,870,044 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -19.61 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16869951 93 - 22407834 UUACAAGACGUUGACAGUUGGCCAACAAACAG-------CAAACGGCCUUGCCGAAGGCCAAUCAAACGUCAUUAUUUCGUUAUACCCAUGUGUG---GUGUG ......((((((....(((((((..(.....)-------....((((...))))..)))))))..))))))..........(((((((....).)---))))) ( -26.50) >DroSec_CAF1 9379 92 - 1 UUACAAGACGUUGACAGUUGGCCAA----CAG-------CAAACGGCCUUGCCGAAGGCCAAUCAAACGUCAUUAUUUCGCUUUACCCAUGUGUGGUGGUGUG ......((((((....(((((((..----(.(-------(((......)))).)..)))))))..)))))).......(((.((((((....).))))).))) ( -29.30) >DroSim_CAF1 11815 92 - 1 UUACAAGACGUUGACAGUUGGCCAA----CAG-------CAAACGGCCUUGCCGAAGGCCAAUCAAACGUCAUUAUUUCGCUUUACCCAUGUGUGGUGGUGUG ......((((((....(((((((..----(.(-------(((......)))).)..)))))))..)))))).......(((.((((((....).))))).))) ( -29.30) >DroEre_CAF1 41610 83 - 1 UUACAAGACGUUGACAGUUGGCCAA----CAG-------CAAACGGCCUUGCCGAAGGCCAAUCAAACGUCAUUAUUUCGUU-UGGCCAUGUG--------UG .............(((..(((((..----...-------.....((((((....))))))....(((((.........))))-))))))..))--------). ( -24.20) >DroYak_CAF1 4708 83 - 1 UUACAAGACGUUGACAGUUGGCCAA----CAG-------CAAACGGCCUUGCCGAAGGCCAAUCAAACGUCAUUAUUUCGUU-UUCCCAUGUG--------UG ......((((((....(((((((..----(.(-------(((......)))).)..)))))))..))))))...........-..........--------.. ( -23.70) >DroPer_CAF1 34445 93 - 1 UUACAAGACGUUGACAUUUGGCCAA----CCGACAGGCUAAAACGGCCUUGCCAAAGGCCAAUCAAACGUCAUUAUCUGGUUAU------AUAUUGUAGUAUA ((((((((((((((..(((((((..----......)))))))..((((((....))))))..)).))))))..((......)).------...)))))).... ( -27.80) >consensus UUACAAGACGUUGACAGUUGGCCAA____CAG_______CAAACGGCCUUGCCGAAGGCCAAUCAAACGUCAUUAUUUCGUU_UACCCAUGUGUG___GUGUG ......((((((....(((((((........((.....))...((((...))))..)))))))..))))))................................ (-19.61 = -20.32 + 0.71)
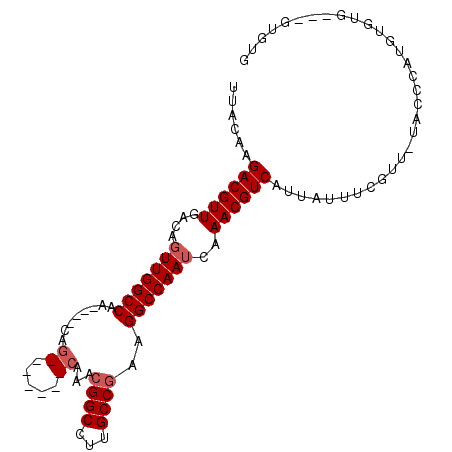
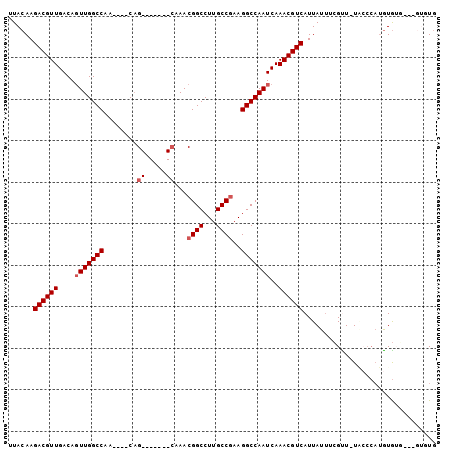
| Location | 16,869,985 – 16,870,075 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -13.45 |
| Energy contribution | -13.95 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
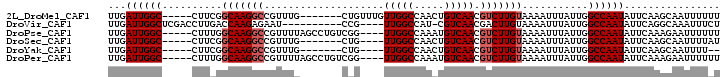
>2L_DroMel_CAF1 16869985 90 + 22407834 UUGAUUGGC-----CUUCGGCAAGGCCGUUUG-------CUGUUUGUUGGCCAACUGUCAACGUCUUGUAAAAUUUAUUGGCCAAUAUUCAAGCAAUUUUUU ......(((-----(((....))))))..(((-------(((..((((((((((.......................))))))))))..).)))))...... ( -27.40) >DroVir_CAF1 23181 87 + 1 UUGAUUGGCUCGACCUUGACCAAGAGAAU----------CCG----UUGGCCAU-CGUCAACGACUUGUAAAAUUUAUUGGCCAAUAUUCAGGCAAAUUUCU ....((((.(((....)))))))((((..----------.((----(((((...-.))))))).................(((........)))...)))). ( -18.10) >DroPse_CAF1 32317 93 + 1 UUGAUUGGC-----CUUUGGCAAGGCCGUUUUAGCCUGUCGG----UUGGCCAAAUGUCAACGUCUUGUAAAAUUUAUUGGCCAAUAUUCAAAGAAUUUUUU ...((((((-----(..(((((.(((.......))))))))(----(((((.....)))))).................)))))))................ ( -25.50) >DroSec_CAF1 9416 86 + 1 UUGAUUGGC-----CUUCGGCAAGGCCGUUUG-------CUG----UUGGCCAACUGUCAACGUCUUGUAAAAUUUAUUGGCCAAUAUUCAAGCAAUUUUAU ......(((-----(((....))))))(((((-------.((----((((((((.......................))))))))))..)))))........ ( -25.40) >DroYak_CAF1 4736 84 + 1 UUGAUUGGC-----CUUCGGCAAGGCCGUUUG-------CUG----UUGGCCAACUGUCAACGUCUUGUAAAAUUUAUUGGCCAAUAUUCAAGCAAUUUU-- ......(((-----(((....))))))(((((-------.((----((((((((.......................))))))))))..)))))......-- ( -25.40) >DroPer_CAF1 34476 93 + 1 UUGAUUGGC-----CUUUGGCAAGGCCGUUUUAGCCUGUCGG----UUGGCCAAAUGUCAACGUCUUGUAAAAUUUAUUGGCCAAUAUUCAAAGAAUUUUUU ...((((((-----(..(((((.(((.......))))))))(----(((((.....)))))).................)))))))................ ( -25.50) >consensus UUGAUUGGC_____CUUCGGCAAGGCCGUUUG_______CUG____UUGGCCAACUGUCAACGUCUUGUAAAAUUUAUUGGCCAAUAUUCAAGCAAUUUUUU ...((((((..........(((((((....................(((((.....))))).)))))))...........))))))................ (-13.45 = -13.95 + 0.50)


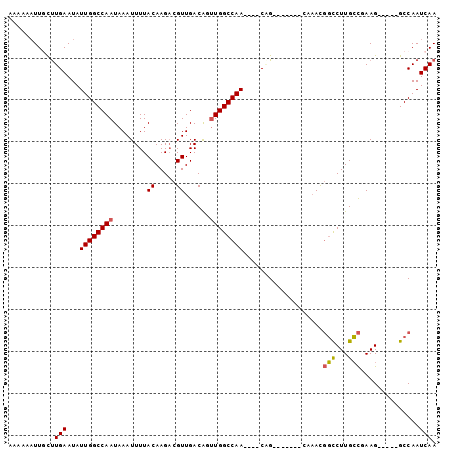
| Location | 16,869,985 – 16,870,075 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.19 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16869985 90 - 22407834 AAAAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAACAAACAG-------CAAACGGCCUUGCCGAAG-----GCCAAUCAA ......(((((......(((((((((.......((.....)).....))))))))).....))-------)))..((((((....)))-----)))...... ( -25.80) >DroVir_CAF1 23181 87 - 1 AGAAAUUUGCCUGAAUAUUGGCCAAUAAAUUUUACAAGUCGUUGACG-AUGGCCAA----CGG----------AUUCUCUUGGUCAAGGUCGAGCCAAUCAA .((..((.(((.((((.((((((((......))....((((....))-))))))))----...----------))))....))).))..))........... ( -18.70) >DroPse_CAF1 32317 93 - 1 AAAAAAUUCUUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAUUUGGCCAA----CCGACAGGCUAAAACGGCCUUGCCAAAG-----GCCAAUCAA ..........((((...((((((((........((.....))......))))))))----((....)).......((((((....)))-----)))..)))) ( -22.84) >DroSec_CAF1 9416 86 - 1 AUAAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAA----CAG-------CAAACGGCCUUGCCGAAG-----GCCAAUCAA ......(((((.(....(((((((((.......((.....)).....)))))))))----)))-------)))..((((((....)))-----)))...... ( -24.50) >DroYak_CAF1 4736 84 - 1 --AAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAA----CAG-------CAAACGGCCUUGCCGAAG-----GCCAAUCAA --....(((((.(....(((((((((.......((.....)).....)))))))))----)))-------)))..((((((....)))-----)))...... ( -24.50) >DroPer_CAF1 34476 93 - 1 AAAAAAUUCUUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAUUUGGCCAA----CCGACAGGCUAAAACGGCCUUGCCAAAG-----GCCAAUCAA ..........((((...((((((((........((.....))......))))))))----((....)).......((((((....)))-----)))..)))) ( -22.84) >consensus AAAAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAA____CAG_______CAAACGGCCUUGCCGAAG_____GCCAAUCAA ...........(((...((((((((........((.....))......))))))))...................(((...)))..............))). (-12.27 = -12.19 + -0.08)

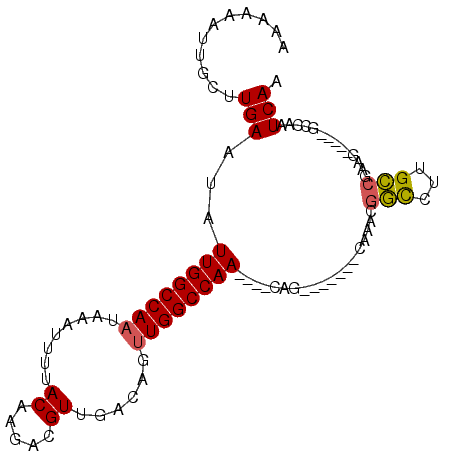
| Location | 16,870,012 – 16,870,103 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -11.31 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16870012 91 - 22407834 AAAAAGCGAUGAACUCGAA--------AUGAAAAAAAAAAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAACAAACAG------- ...(((((((.........--------..............))))))).....(((((((((.......((.....)).....))))))))).......------- ( -16.10) >DroPse_CAF1 32344 100 - 1 AAAAAGAGA--GACUGAGAGGGAGACAUUCAAAAACAAAAAAUUCUUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAUUUGGCCAA----CCGACAGGCU .........--..(((...((.....(((((((............))))))).((((((((........((.....))......))))))))----))..)))... ( -18.44) >DroSec_CAF1 9443 87 - 1 AAAAAGCGAUGAACUCAAA--------AUGAAAAAAAUAAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAA----CAG------- ...(((((((.........--------..............))))))).....(((((((((.......((.....)).....)))))))))----...------- ( -16.10) >DroEre_CAF1 41665 82 - 1 AAAAAGCGAUGAACUGAAC--------AUAAAA-----AAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAA----CAG------- ...(((((((.........--------......-----...))))))).....(((((((((.......((.....)).....)))))))))----...------- ( -16.37) >DroYak_CAF1 4763 83 - 1 AAAAAGCAAUGAACUUAAA--------AUAAAAA----AAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAA----CAG------- ...(((((((.........--------.......----...))))))).....(((((((((.......((.....)).....)))))))))----...------- ( -16.61) >DroPer_CAF1 34503 100 - 1 AAAAAGAGA--GACUGAGAGGGAGACAUUCAAAAACAAAAAAUUCUUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAUUUGGCCAA----CCGACAGGCU .........--..(((...((.....(((((((............))))))).((((((((........((.....))......))))))))----))..)))... ( -18.44) >consensus AAAAAGCGAUGAACUGAAA________AUAAAAAA_AAAAAAUUGCUUGAAUAUUGGCCAAUAAAUUUUACAAGACGUUGACAGUUGGCCAA____CAG_______ ...(((((((...............................))))))).....(((((((((.......((.....)).....))))))))).............. (-11.31 = -11.95 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:16 2006