| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,868,213 – 16,868,303 |
| Length | 90 |
| Max. P | 0.957645 |

| Location | 16,868,213 – 16,868,303 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -18.06 |
| Energy contribution | -18.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16868213 90 + 22407834 UUUUUUCUCGCGCCAGGAAAUUGGCAACUAAGCCAAGCCAAGCCAAGUCCAAGGCAUUGUGUCCUUCCUUUUCCUCUUGACUGGCUGCAU .........(((((((..(((((((......)))))((((((((........))).))).))..............))..))))).)).. ( -24.50) >DroSec_CAF1 7630 86 + 1 U--UUUCU--CGCCAGGAAAUUGGCAACUAAGCCAAGCCAAGCCAAGUCCAAGGCAUUGUGUCCUUCCUUUUCCUCUUGACUGGCUGCAU .--.....--.((..((...(((((......))))).)).(((((.(((..(((.(..(........)..).)))...)))))))))).. ( -22.30) >DroSim_CAF1 10100 83 + 1 UUUUUUCU--CGCCAGGAAAUUGGCAACUAAGCCA-----AGCCAAGUCCAAAGCAUUGUGUCCUUCCUUUUCCUCUUGACUGGCUGCAU ........--.(((((....(((((......))))-----)..((((...((((.............))))....)))).)))))..... ( -17.62) >DroEre_CAF1 39886 80 + 1 U---UUCU--CGCCAGGAAAUUGGCAACUAAGCCA-----AGCCAAGUCCAAGGCAUUGUGUCCUUCCUUUUCCUCUUGACUGGCUGCAU .---....--.(((((..(((((((......))))-----)(((........))).....................))..)))))..... ( -20.70) >DroYak_CAF1 2993 80 + 1 U---UUCU--CGCCAGGAAAUUGGCAACUAAGCCA-----AGCCAAGUCCAAGGCAUUGUGUCCUUUCUUUUCCUCUUGACUGGCUGCAU .---....--.(((((..(((((((......))))-----)(((........))).....................))..)))))..... ( -20.70) >consensus U__UUUCU__CGCCAGGAAAUUGGCAACUAAGCCA_____AGCCAAGUCCAAGGCAUUGUGUCCUUCCUUUUCCUCUUGACUGGCUGCAU ...........(((((..((.((((......))))......(((........))).....................))..)))))..... (-18.06 = -18.26 + 0.20)



| Location | 16,868,213 – 16,868,303 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16868213 90 - 22407834 AUGCAGCCAGUCAAGAGGAAAAGGAAGGACACAAUGCCUUGGACUUGGCUUGGCUUGGCUUAGUUGCCAAUUUCCUGGCGCGAGAAAAAA .((((((((((((((.....(((.((((.(.....)))))...)))..)))))).))))).....((((......)))))))........ ( -27.20) >DroSec_CAF1 7630 86 - 1 AUGCAGCCAGUCAAGAGGAAAAGGAAGGACACAAUGCCUUGGACUUGGCUUGGCUUGGCUUAGUUGCCAAUUUCCUGGCG--AGAAA--A ....(((((((((((.....(((.((((.(.....)))))...)))..)))))).)))))...((((((......)))))--)....--. ( -25.90) >DroSim_CAF1 10100 83 - 1 AUGCAGCCAGUCAAGAGGAAAAGGAAGGACACAAUGCUUUGGACUUGGCU-----UGGCUUAGUUGCCAAUUUCCUGGCG--AGAAAAAA ....(((((((((((....((((.(.........).))))...)))))).-----)))))...((((((......)))))--)....... ( -21.80) >DroEre_CAF1 39886 80 - 1 AUGCAGCCAGUCAAGAGGAAAAGGAAGGACACAAUGCCUUGGACUUGGCU-----UGGCUUAGUUGCCAAUUUCCUGGCG--AGAA---A ....(((((((((((.....((((..(....)....))))...)))))).-----)))))...((((((......)))))--)...---. ( -23.60) >DroYak_CAF1 2993 80 - 1 AUGCAGCCAGUCAAGAGGAAAAGAAAGGACACAAUGCCUUGGACUUGGCU-----UGGCUUAGUUGCCAAUUUCCUGGCG--AGAA---A ....(((((((((((.........((((.(.....)))))...)))))).-----)))))...((((((......)))))--)...---. ( -22.10) >consensus AUGCAGCCAGUCAAGAGGAAAAGGAAGGACACAAUGCCUUGGACUUGGCU_____UGGCUUAGUUGCCAAUUUCCUGGCG__AGAAA__A .....(((((..((..((......((((.(.....))))).((((.((((......)))).)))).))..))..)))))........... (-21.32 = -21.16 + -0.16)

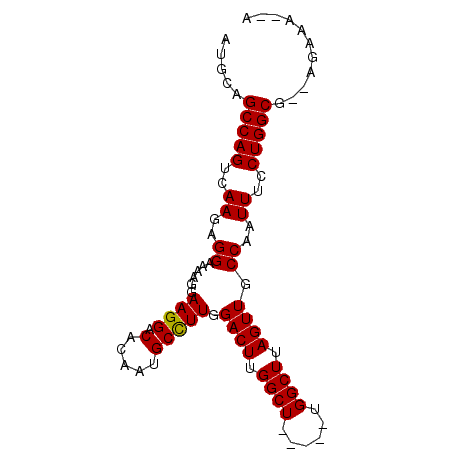

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:11 2006