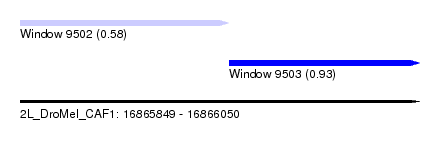
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,865,849 – 16,866,050 |
| Length | 201 |
| Max. P | 0.926661 |

| Location | 16,865,849 – 16,865,954 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -32.88 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
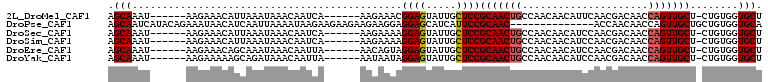
>2L_DroMel_CAF1 16865849 105 + 22407834 CACACAUUGCCUUAUGGCCACUGGACGAUGGACCACAGUGGAGCCUGGGCAGUACGCAACACCUGACGCUGCCCGCCAGGAUACUCCCGUUCGUCUCGAUCAUAC ....((..(((....)))...))((.(((((((...((((...((((((((((...((.....))..)))))))...))).))))...)))))))))........ ( -31.50) >DroSec_CAF1 5370 105 + 1 CACACAGUGCCUUAUGGCCACUGGACGAUGGACCACAGUGGAGCCUGGGCAGUAUGCAACACCUGACGCUGCCCGCCAGGAUACUCCCGUUCGUCUCGAUCAUAC ....(((((((....)).)))))((.(((((((...((((...((((((((((...((.....))..)))))))...))).))))...)))))))))........ ( -37.10) >DroSim_CAF1 7819 105 + 1 CACACAGUGCCUUAUAGCCACUGGACGAUGGACCACAGUGGAGCCUGGGCAGUACGCAACACCUGACGCUGCCCGCCAGGAUACUCCCGUUCGUCUCGAUCAUAC ....(((((.(.....).)))))((.(((((((...((((...((((((((((...((.....))..)))))))...))).))))...)))))))))........ ( -33.50) >DroEre_CAF1 37642 105 + 1 CACACAGUGCCUUAUGGCCACUGGAUGAAGGACCACAGUGGAGCCUGGGCAGUACGCAACACCUGACGCUGCCCGCCAGGAUACUCCCGUCCGUCUCGAUCAUAC ....(((((((....)).)))))((.((.((((...((((...((((((((((...((.....))..)))))))...))).))))...)))).))))........ ( -36.10) >DroYak_CAF1 552 104 + 1 CACACAGUGCUUUAUGGCCACUGCACGAUGGACCACAGUGGAGCCUGGGCAGUACGCAACACCUGACACUGCUCGCC-GGAUACUCCCGUCCGUCCCGAUCAUAC ....(((((((....)).)))))...(((((((...((((...((((((((((...((.....))..))))))))..-)).))))...))))))).......... ( -31.80) >consensus CACACAGUGCCUUAUGGCCACUGGACGAUGGACCACAGUGGAGCCUGGGCAGUACGCAACACCUGACGCUGCCCGCCAGGAUACUCCCGUUCGUCUCGAUCAUAC ....(((((((....)).)))))...(((((((...((((...((((((((((...((.....))..)))))))...))).))))...))))))).......... (-32.88 = -33.00 + 0.12)


| Location | 16,865,954 – 16,866,050 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -15.29 |
| Energy contribution | -15.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16865954 96 + 22407834 AGCAAAU------AAGAAACAUUAAAUAAACAAUCA------AAGAAACGGAGUAUUGCUCCGCAACUGCCAACAACAUUCAACGACAACCAGUUGCU-CUGUGGUGCU ((((...------.(((...................------.......((((.....))))(((((((.....................))))))))-))....)))) ( -20.00) >DroPse_CAF1 27872 95 + 1 AGCAAUCAUACAGAAAUAACAUCAAUUAAAAUAAGAAGAAGAAGAAGGAGGAGCAUCAUUCCGCAAC--------------ACCAACAACCAGUUGCUGCUGUGGUGCA .(((....(((((.................................((((........))))(((((--------------...........)))))..))))).))). ( -17.50) >DroSec_CAF1 5475 96 + 1 AGCAAAU------AAGAAACAUUAAAUAAACAAUCA------AAGAAAAGGAGUAUUGCUCCGCAACUGCCAACAACAUCCAACGACAACCAGUUGCU-CUGUGGUGCU ((((...------.(((...................------.......((((.....))))(((((((.....................))))))))-))....)))) ( -19.90) >DroSim_CAF1 7924 96 + 1 AGCAAAU------AAGAAACAUUAAAUAAACAAUCA------AAGAAAAGGAGUAUUGCUCCGCAACUGCCAACAACAUCCAACGACAACCAGUUGCU-CUGUGGUGCU ((((...------.(((...................------.......((((.....))))(((((((.....................))))))))-))....)))) ( -19.90) >DroEre_CAF1 37747 96 + 1 AGCAAAU------AAGAAACAGCAAAUAAACAAUUA------AACAGUAGGAGUAUUGCUCCGCAACUGCCAACAACAUCCAACGACAACCAGUUGCU-CUGUGGUGCU ((((...------.(((...................------.......((((.....))))(((((((.....................))))))))-))....)))) ( -19.90) >DroYak_CAF1 656 96 + 1 AGCAAAU------AAGAAAAAGCAGAUAAACAAUUA------AAUAAUAGGAGUAUUGCUCCGCAACUGCCAACAACAUCCAACGACAACCAGUUGCU-CUGUGGUGCU ((((...------........(((((..........------.......((((.....))))(((((((.....................))))))))-))))..)))) ( -20.82) >consensus AGCAAAU______AAGAAACAUCAAAUAAACAAUCA______AAGAAAAGGAGUAUUGCUCCGCAACUGCCAACAACAUCCAACGACAACCAGUUGCU_CUGUGGUGCU .(((.............................................((((.....))))(((((((.....................)))))))........))). (-15.29 = -15.48 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:06 2006