
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,841,322 – 16,841,416 |
| Length | 94 |
| Max. P | 0.985929 |

| Location | 16,841,322 – 16,841,416 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.59 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -29.27 |
| Energy contribution | -29.93 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
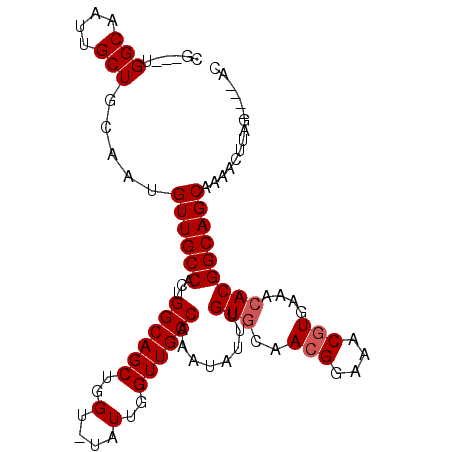
>2L_DroMel_CAF1 16841322 94 + 22407834 AG---UGGCAAUUGCUGCAAUGUUGCCACAGGCAGCUGGUUUAUUGGUUGCCAAAUAUUUGUGCAACGGAAACGUGAAACACGGCAGCAAAACUUGG------ .(---..((....))..)..(((((((...((((((..(....)..))))))........(((..(((....)))....))))))))))........------ ( -37.70) >DroEre_CAF1 943 102 + 1 CGUGGUGGCAAUUGCUGCAAUGUUGCCACUGGCAGCUGGU-UAUUGGUUGCCAAAUAUUUGUGCAACGGAAACGUGAAGCACGGCAGCCAAACUUAGGAGUAC ....(..((....))..)...((((((..(((((((..(.-..)..))))))).......((((.(((....)))...))))))))))............... ( -39.00) >DroAna_CAF1 650 98 + 1 CG---UGGCAAUUGCUGCAAUGUUGCCACUGGCAGCUGGU-UAUUGGUUGCCAAAUAUUUGUGCAACGGAAACAUGAAA-ACGGCAGCAACACCACAAUACAC .(---(((...(((((((...(((((((.(((((((..(.-..)..)))))))......)).)))))(....)......-...)))))))..))))....... ( -36.60) >consensus CG___UGGCAAUUGCUGCAAUGUUGCCACUGGCAGCUGGU_UAUUGGUUGCCAAAUAUUUGUGCAACGGAAACGUGAAACACGGCAGCAAAACUUAG____AC ......(((....))).....((((((...((((((..(....)..))))))........(((..(((....)))....)))))))))............... (-29.27 = -29.93 + 0.67)

| Location | 16,841,322 – 16,841,416 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.59 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16841322 94 - 22407834 ------CCAAGUUUUGCUGCCGUGUUUCACGUUUCCGUUGCACAAAUAUUUGGCAACCAAUAAACCAGCUGCCUGUGGCAACAUUGCAGCAAUUGCCA---CU ------....((.(((((((.(((((....((((..(((((.(((....))))))))....))))..((((....))))))))).)))))))..))..---.. ( -25.80) >DroEre_CAF1 943 102 - 1 GUACUCCUAAGUUUGGCUGCCGUGCUUCACGUUUCCGUUGCACAAAUAUUUGGCAACCAAUA-ACCAGCUGCCAGUGGCAACAUUGCAGCAAUUGCCACCACG ..........((.((((((((((((...(((....))).))))........)))).......-....(((((.((((....)))))))))....))))..)). ( -29.90) >DroAna_CAF1 650 98 - 1 GUGUAUUGUGGUGUUGCUGCCGU-UUUCAUGUUUCCGUUGCACAAAUAUUUGGCAACCAAUA-ACCAGCUGCCAGUGGCAACAUUGCAGCAAUUGCCA---CG .......(((((((((((((...-......(((...(((((.(((....))))))))....)-))..(.((((...)))).)...))))))).)))))---). ( -30.20) >consensus GU____CCAAGUUUUGCUGCCGUGUUUCACGUUUCCGUUGCACAAAUAUUUGGCAACCAAUA_ACCAGCUGCCAGUGGCAACAUUGCAGCAAUUGCCA___CG ...............((...................(((((.(((....))))))))..........(((((.((((....)))))))))....))....... (-21.07 = -21.40 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:56 2006