| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,840,588 – 16,840,700 |
| Length | 112 |
| Max. P | 0.799099 |

| Location | 16,840,588 – 16,840,700 |
|---|---|
| Length | 112 |
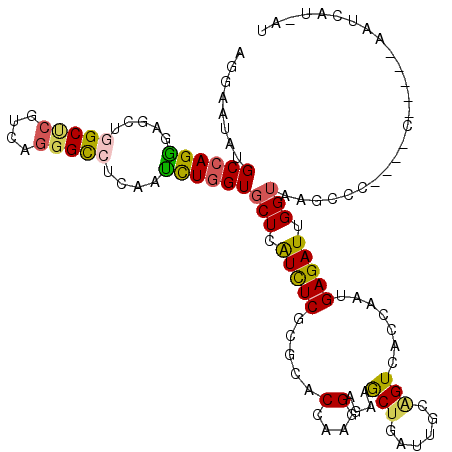
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.32 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16840588 112 - 22407834 AGGAGUACGCCAGGGAGCUGGCUCGCCAGGGUAUCAACCUGGUGCUCAUCUCGCGGACGAAGGAGAAGCUCAUUGCGGUUACCAAUGAGAUUGGUAAGUGU--UGC----UUAGUA--CU ...((((((((((....))))).(((((((.......)))))))(((...(((....)))..)))((((.....(((.((((((((...)))))))).)))--.))----)).)))--)) ( -43.40) >DroPse_CAF1 1264 120 - 1 AGGAAUAUGCCAGAGAGUUGGCCCGUCAGGGCCUUAAUCUGGUGCUUGUCUCUCGCACCAAGGAGAAGCUGAUUGCAGUCACCAAUGAGAUUGGUAAGCUCAAACCCAGUCAAUCUUUAU .((.....((((((.....(((((....)))))....))))))((.........)).))(((((...((((.(((.(((.((((((...))))))..))))))...))))...))))).. ( -36.50) >DroEre_CAF1 153 114 - 1 AGGAGUAUGCCAGGGAGCUGGCUCGACAGGGUAUCAACCUGGUGCUCAUCUCGCGGACGAAGGAGAAGCUUAUUGCGGUGAGCAAUGAGAUUGGUAAGCAU--GUC----UAAGCAUGAU ..(((((((((((....)))))....((((.......)))))))))).((((.(.......)))))..((((((((.....)))))))).........(((--((.----...))))).. ( -39.10) >DroWil_CAF1 173 109 - 1 AGGAAUAUGCCAGGGAGCUGGCACGUCAAGGCCUCAAUCUGGUUCUGAUUUCACGUACCAAGGAGAAGCUGAUCCAAGUGACCAAUGAGAUAGGUAAAAC-----UCU---GAUCUC--- .((....((((((....)))))).((((..(((.......)))..))))........))...........((((..(((.(((.........)))...))-----)..---))))..--- ( -29.20) >DroAna_CAF1 156 104 - 1 AGGAAUAUGCCAGGGAACUGGCUCGCCAGGGCCUCAAUUUAGUGCUCAUCUCGCGCACCAAGGAGAAACUCAUAGCUGUUACCAACGAGAUUGGUAAGUCC--------------A--CA .((.....(((((....)))))...)).(((((((......((((.(.....).))))....))).............(((((((.....)))))))))))--------------.--.. ( -31.30) >DroPer_CAF1 1294 120 - 1 AGGAAUAUGCCAGAGAGUUGGCCCGUCAGGGCCUUAAUCUGGUGCUUGUCUCUCGCACCAAGGAGAAGCUGAUUGCAGUCACCAAUGAGAUUGGUAAGCUCAAACCCAGUCAAUCUUUAU .((.....((((((.....(((((....)))))....))))))((.........)).))(((((...((((.(((.(((.((((((...))))))..))))))...))))...))))).. ( -36.50) >consensus AGGAAUAUGCCAGGGAGCUGGCUCGUCAGGGCCUCAAUCUGGUGCUCAUCUCGCGCACCAAGGAGAAGCUGAUUGCAGUCACCAAUGAGAUUGGUAAGCCC____C_____AAUCAU_AU ........((((((.....(((((....)))))....))))))(((.(((((.....(....)....(((......))).......))))).)))......................... (-21.78 = -21.32 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:54 2006