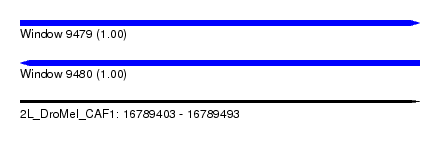
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,789,403 – 16,789,493 |
| Length | 90 |
| Max. P | 0.999947 |

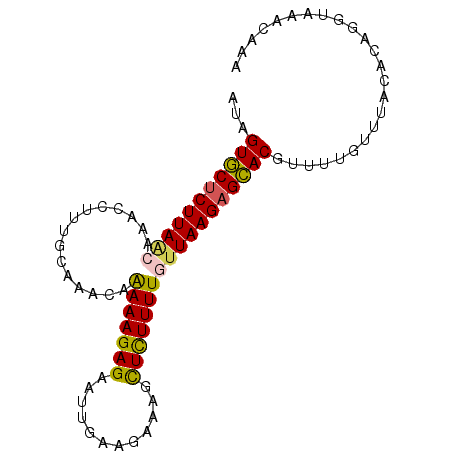
| Location | 16,789,403 – 16,789,493 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -11.03 |
| Energy contribution | -14.66 |
| Covariance contribution | 3.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16789403 90 + 22407834 UUUGUUUACCUGUCUAAACAAAACGUGCUCUUAACAAAAGAGCUUUCUUCGAUUCUCUUUUUGUUUGCAAAGGUUUGUUAAGAGCACUAU ((((((((......))))))))..((((((((((((((((((..((....))..))))(((((....))))).))))))))))))))... ( -28.30) >DroSec_CAF1 9518 90 + 1 UUUGUUUACCUGUGUAAACAAAACGUGCUCUUAACAAAAGAGCUUUCUUCAAUUCUCUUUUUGUUUGCAAAGGUUGCUUAAGAGCACUAU (((((((((....)))))))))..((((((((((.(((((((..((....))..))))))).....(((.....)))))))))))))... ( -26.50) >DroSim_CAF1 9617 90 + 1 UUUGUUUACCUGUGUAAAUAAAACGUGCUCUUAACAAAAGAGCUUUCCUCUAUUCUCUUUUUGUUUGGAAAUGAUGCCUAAGAGCACUAU (((((((((....)))))))))..((((((((((((((((((..(......)..))).))))))..((........)))))))))))... ( -23.30) >DroYak_CAF1 11835 72 + 1 ----------UGUCUACGCAAUACGUUCACUUAUCAAAAAAACUUUGUUCAAUUCUCUUUCUCUUU--------UUGUUAAGAGAACUUU ----------..............((((.((((.((((((.......................)))--------))).)))).))))... ( -8.70) >consensus UUUGUUUACCUGUCUAAACAAAACGUGCUCUUAACAAAAGAGCUUUCUUCAAUUCUCUUUUUGUUUGCAAAGGUUGCUUAAGAGCACUAU ((((((((......))))))))..((((((((((((((((((..((....))..)))))))...............)))))))))))... (-11.03 = -14.66 + 3.63)

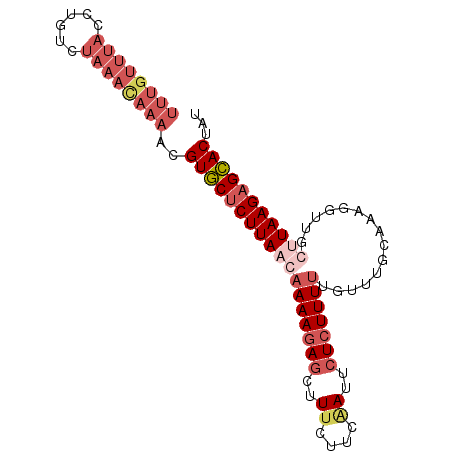
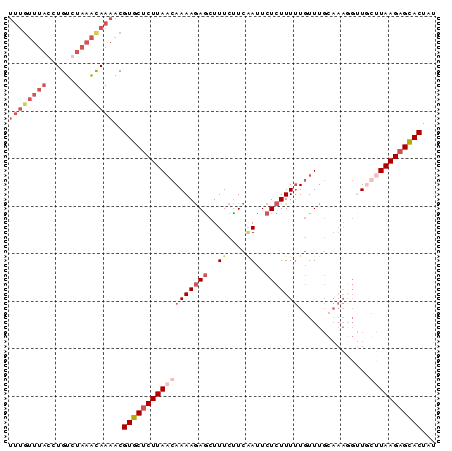
| Location | 16,789,403 – 16,789,493 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.51 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.76 |
| SVM RNA-class probability | 0.999947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16789403 90 - 22407834 AUAGUGCUCUUAACAAACCUUUGCAAACAAAAAGAGAAUCGAAGAAAGCUCUUUUGUUAAGAGCACGUUUUGUUUAGACAGGUAAACAAA ...(((((((((((((...((((....))))(((((..((...))...))))))))))))))))))..((((((((......)))))))) ( -26.10) >DroSec_CAF1 9518 90 - 1 AUAGUGCUCUUAAGCAACCUUUGCAAACAAAAAGAGAAUUGAAGAAAGCUCUUUUGUUAAGAGCACGUUUUGUUUACACAGGUAAACAAA ...(((((((((((((.....))).....(((((((..((....))..))))))).))))))))))..(((((((((....))))))))) ( -29.30) >DroSim_CAF1 9617 90 - 1 AUAGUGCUCUUAGGCAUCAUUUCCAAACAAAAAGAGAAUAGAGGAAAGCUCUUUUGUUAAGAGCACGUUUUAUUUACACAGGUAAACAAA ...(((((((((((........)).....(((((((............)))))))..)))))))))......(((((....))))).... ( -20.20) >DroYak_CAF1 11835 72 - 1 AAAGUUCUCUUAACAA--------AAAGAGAAAGAGAAUUGAACAAAGUUUUUUUGAUAAGUGAACGUAUUGCGUAGACA---------- .....((((((.....--------.))))))(((((((((......))))))))).....((..(((.....)))..)).---------- ( -12.80) >consensus AUAGUGCUCUUAACAAACCUUUGCAAACAAAAAGAGAAUUGAAGAAAGCUCUUUUGUUAAGAGCACGUUUUGUUUACACAGGUAAACAAA ...(((((((((((...............(((((((............))))))))))))))))))........................ (-14.57 = -14.51 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:44 2006