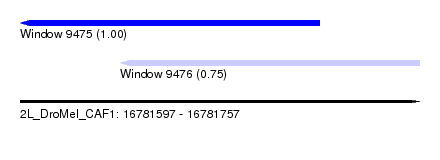
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,781,597 – 16,781,757 |
| Length | 160 |
| Max. P | 0.995502 |

| Location | 16,781,597 – 16,781,717 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -47.93 |
| Consensus MFE | -44.02 |
| Energy contribution | -44.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
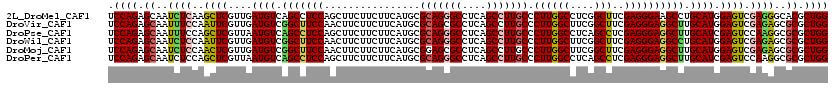
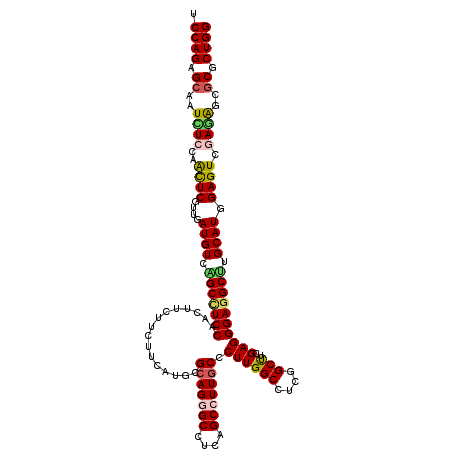
>2L_DroMel_CAF1 16781597 120 - 22407834 UCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCGGCUUCGAGGGAAGCCUGCAUGGAGUCGAGGGCACGCUGGUGGUUCUUGCGGGUGUUCUCGAACUCCUCCUCCUUCUCCU ((.....(((.((((((((((((((..((((((((((..(((((((....)))))).....)..)))))))))).))))).)).))))))).))).....)).................. ( -50.40) >DroPse_CAF1 4695 120 - 1 UCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCAGCCUCGAGGGAGGCUUGCAUCGAGUCCAAGGCGCGCUGGUGGUUCUUGCGGGUGUUCUCGAACUCCUCUUCCUUCUCCU ((.....(((.((((((((((((((...((((((((.(((((((((....))))))......))).)))))).))))))).)).))))))).))).....)).................. ( -46.80) >DroSim_CAF1 1834 120 - 1 UCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCGGCUUCGAGGGAAGCCUGCAUGGAGUCGAGGGCACGCUGGUGGUUCUUGCGGGUGUUCUCGAACUCCUCCUCCUUCUCCU ((.....(((.((((((((((((((..((((((((((..(((((((....)))))).....)..)))))))))).))))).)).))))))).))).....)).................. ( -50.40) >DroYak_CAF1 4082 120 - 1 UCUUCUUCAUGCGCAGGGCCUCAGCCUUACCCUUGGCCUCGGCUUCAAGGGAAGCCUGCAUGGAGUCGAGGGCACGCUGGUGGUUCUUGCGGGUGUUCUCGAACUCCUCCUCCUUCUCCU ....(((((((((((((((....))))).((((((((....))..))))))..))..))))))))((((((((((.(((..........))))))))))))).................. ( -45.20) >DroAna_CAF1 1829 120 - 1 UCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCAGCCUCAAGGGAAGCCUGCAUGGAGUCGAGGGCGCGCUGGUGGUUCUUGCGGGUGUUCUCGAACUCCUCCUCCUUCUCCU ((.....(((.((((((((((((((..((((((((((..((((....(((....))))).))..)))))))))).))))).)).))))))).))).....)).................. ( -48.00) >DroPer_CAF1 4567 120 - 1 UCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCAGCCUCGAGGGAGGCUUGCAUCGAGUCCAAGGCGCGCUGGUGGUUCUUGCGGGUGUUCUCGAACUCCUCUUCCUUCUCCU ((.....(((.((((((((((((((...((((((((.(((((((((....))))))......))).)))))).))))))).)).))))))).))).....)).................. ( -46.80) >consensus UCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCAGCCUCGAGGGAAGCCUGCAUGGAGUCGAGGGCACGCUGGUGGUUCUUGCGGGUGUUCUCGAACUCCUCCUCCUUCUCCU ((.....(((.((((((((((((((..((((((((((.((((((((....))))))......)))))))))))).))))).)).))))))).))).....)).................. (-44.02 = -44.27 + 0.25)


| Location | 16,781,637 – 16,781,757 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -47.17 |
| Consensus MFE | -40.68 |
| Energy contribution | -40.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16781637 120 - 22407834 UCCAGAGCAAUCUCAAGCUCGUUGAUGUCAGCCUCCAGCUUCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCGGCUUCGAGGGAAGCCUGCAUGGAGUCGAGGGCACGCUGG ....((((........))))((((....))))..(((((.............(((((((....)))))))...((.((((((((((((((....)))...))))))))))).)).))))) ( -51.20) >DroVir_CAF1 1915 120 - 1 UCCAGAGCAAUUUCCAAUUCGUUGAUGUCGGCUUCCAACUUCUUCUUCAUGCGCAGCGCCUCAGCCUUGCCCUUGGCUUCGGCUUCGAGGGAGGCUUGCAUGGAGUCGAGAGCGCGCUGG .((((.((............((((....)))).......((((((((((((((....(((((..(((((.....(((....))).)))))))))).)))))))))..))))).)).)))) ( -42.30) >DroPse_CAF1 4735 120 - 1 UCCAGAGCAAUUUCCAGCUCGUUAAUGUCAGCCUCCAGCUUCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCAGCCUCGAGGGAGGCUUGCAUCGAGUCCAAGGCGCGCUGG .((((.((..(((...(((((...((((.(((((((.((...........))(((((((....))))))).((((((....)))..)))))))))).)))))))))..)))..)).)))) ( -46.70) >DroWil_CAF1 1331 120 - 1 UCCAGAGCAAUCUCCAAUUCGUUGAUGUCGGCUUCCAACUUCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCUUCGGCUUCGAGGGAGGCCUGCAUGGAGUCGAGAGCGCGCUGG .((((.((..((((......((((....))))............((((((((...(((((((..(((((.....(((....))).))))))))))))))))))))..))))..)).)))) ( -49.00) >DroMoj_CAF1 1898 120 - 1 UCCAGAGCAAUCUCCAACUCGUUGAUGUCGGCUUCCAACUUCUUCUUCAUGCGGAGCGCCUCAGCCUUGCCCUUGGCUUCGGCUUCGAGGGAGGCUUGCAUGGAGUCGAGAGCGCGCUGG .((((.((..((((..((((((((....))))...............(((((((((...)))(((((..((((((((....))..))))))))))))))))))))).))))..)).)))) ( -46.70) >DroPer_CAF1 4607 120 - 1 UCCAGAGCAAUCUCCAGCUCGUUAAUGUCAGCCUCCAGCUUCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCAGCCUCGAGGGAGGCUUGCAUCGAGUCCAAGGCGCGCUGG .((((.((..(((...(((((...((((.(((((((.((...........))(((((((....))))))).((((((....)))..)))))))))).)))))))))...))).)).)))) ( -47.10) >consensus UCCAGAGCAAUCUCCAACUCGUUGAUGUCAGCCUCCAACUUCUUCUUCAUGCGCAGGGCCUCAGCCUUGCCCUUGGCCUCGGCUUCGAGGGAGGCUUGCAUGGAGUCGAGAGCGCGCUGG .((((.((..((((..((((....((((.(((((((................(((((((....))))))).((((((....)))..)))))))))).)))).)))).))))..)).)))) (-40.68 = -40.52 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:40 2006