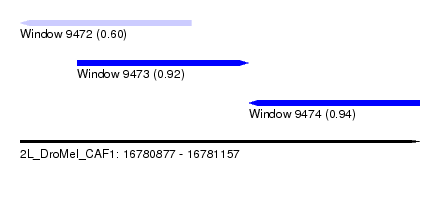
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,780,877 – 16,781,157 |
| Length | 280 |
| Max. P | 0.939668 |

| Location | 16,780,877 – 16,780,997 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.78 |
| Mean single sequence MFE | -42.87 |
| Consensus MFE | -36.92 |
| Energy contribution | -36.40 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
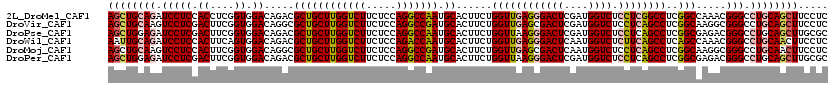
>2L_DroMel_CAF1 16780877 120 - 22407834 ACUCGUACUUGCUGCGCCACACCUGGGCCUCAGCGUUGGCCUUGCUCAGCUGGCGCUGCAGAUCGGCCUUGCCCUCAGCCUCCUCCUCAACCUGCUCGCGCAGAUUGUCCAGGUCGUGCU .............((((...((((((((((((((((..((........))..)))))).))...(((..........)))...........((((....))))...)))))))).)))). ( -42.30) >DroVir_CAF1 1155 120 - 1 ACUCGUACUUGCUGCGCCAGACCUGGGCCUCAGCGUUAGCCUUGCUGAGUUGACGCUGCAAAUCAGCCUUGCCCUCAGCCUCCUCCUCAACCUGCUCGCGCAAGUUGUCGAGGUCGUGCU ....((((..((((.((.((..((((....(((((((((((.....).))))))))))....)))).)).))...))))..((((..((((.(((....))).))))..))))..)))). ( -43.30) >DroPse_CAF1 3975 120 - 1 ACUCGUACUUGCUGCGCCAGAUCUGGGACUCGGCGUUGGCCUUGCUCAAUUGACGUUGCAGAUCAGCCUUGCCCUCGGCCUCCUCCUCAACCUGUUCGCGCAGGUUGUCGAGGUCGUGCU ....((((..((...((..((((((.((((((..((((((...)).))))))).))).)))))).))...))....((((((.....((((((((....))))))))..)))))))))). ( -43.30) >DroGri_CAF1 1263 120 - 1 AUUCGUACUUGCUGCGCCAGACCUGAGCCUCAGCGUUAGCCUUGCUGAGUUGACGCUGCAGAUCAGCCUUGCCCUCAGCCUCCUCCUCAACCUGUUCGCGCAGGUUGUCCAGGUCGUGCU ....((((..((((.((.((..((((..(((((((((((((.....).)))))))))).)).)))).)).))...))))..(((...((((((((....))))))))...)))..)))). ( -45.20) >DroWil_CAF1 571 120 - 1 AUUCGUACUUGCUACGCCAGACCUGGGCUUCGGCGUUGGCCUUGCUCAAUUGACGCUGCAAAUCAGCCUUACCCUCAGCCUCCUCCUCAACCUGUUCGCGCAGGUUGUCGAGGUCGUGCU ....((((..(((..((.((.((.(.((....)).).)).)).)).....(((.((((.....)))).))).....)))..((((..((((((((....))))))))..))))..)))). ( -39.80) >DroPer_CAF1 3847 120 - 1 ACUCGUACUUGCUGCGCCAGAUCUGGGACUCGGCGUUGGCCUUGCUCAAUUGACGUUGCAGAUCAGCCUUGCCCUCGGCCUCCUCCUCAACCUGUUCGCGCAGGUUGUCGAGGUCGUGCU ....((((..((...((..((((((.((((((..((((((...)).))))))).))).)))))).))...))....((((((.....((((((((....))))))))..)))))))))). ( -43.30) >consensus ACUCGUACUUGCUGCGCCAGACCUGGGCCUCAGCGUUGGCCUUGCUCAAUUGACGCUGCAGAUCAGCCUUGCCCUCAGCCUCCUCCUCAACCUGUUCGCGCAGGUUGUCGAGGUCGUGCU ....((((..((((.((.((..((((....(((((((((..........)))))))))....)))).)).))...))))..((((..((((((((....))))))))..))))..)))). (-36.92 = -36.40 + -0.52)

| Location | 16,780,917 – 16,781,037 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -49.30 |
| Consensus MFE | -46.21 |
| Energy contribution | -45.97 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16780917 120 + 22407834 GGCUGAGGGCAAGGCCGAUCUGCAGCGCCAGCUGAGCAAGGCCAACGCUGAGGCCCAGGUGUGGCGCAGCAAGUACGAGUCCGAUGGCGUUGCCCGCUCUGAGGAGCUGGAGGAAGCCAA ((((..((((((.((((...(((.((((((.(((.((..(((....)))...)).)))...)))))).)))....((....)).)))).))))))((((....)))).......)))).. ( -55.20) >DroVir_CAF1 1195 120 + 1 GGCUGAGGGCAAGGCUGAUUUGCAGCGUCAACUCAGCAAGGCUAACGCUGAGGCCCAGGUCUGGCGCAGCAAGUACGAGUCCGAUGGUGUUGCCCGCUCUGAGGAGCUGGAGGAGGCCAA ((((..((((((.(((.((((((.((((((.((((((.........))))))((....)).)))))).)))))).((....))..))).))))))((((....)))).......)))).. ( -52.00) >DroPse_CAF1 4015 120 + 1 GGCCGAGGGCAAGGCUGAUCUGCAACGUCAAUUGAGCAAGGCCAACGCCGAGUCCCAGAUCUGGCGCAGCAAGUACGAGUCCGAUGGUGUUGCCCGCUCUGAGGAGCUGGAGGAGGCCAA ((((..((((((.(((....(((..((((((((..((..(((....)))..))....))).)))))..)))....((....))..))).))))))((((....)))).......)))).. ( -43.90) >DroGri_CAF1 1303 120 + 1 GGCUGAGGGCAAGGCUGAUCUGCAGCGUCAACUCAGCAAGGCUAACGCUGAGGCUCAGGUCUGGCGCAGCAAGUACGAAUCCGAUGGCGUUGCCCGCUCCGAGGAGCUAGAGGAGGCCAA ((((..((((((.(((....(((.((((((.((.(((..(((....)))...))).))...)))))).)))....((....))..))).))))))((((....)))).......)))).. ( -53.00) >DroWil_CAF1 611 120 + 1 GGCUGAGGGUAAGGCUGAUUUGCAGCGUCAAUUGAGCAAGGCCAACGCCGAAGCCCAGGUCUGGCGUAGCAAGUACGAAUCGGACGGUGUUGCCCGUUCCGAGGAGUUGGAGGAAGCCAA ((((..((((((.((((((((((.((((((.(((.((..(((....)))...)).)))...)))))).)))))).((...))..)))).)))))).((((((....))))))..)))).. ( -47.80) >DroPer_CAF1 3887 120 + 1 GGCCGAGGGCAAGGCUGAUCUGCAACGUCAAUUGAGCAAGGCCAACGCCGAGUCCCAGAUCUGGCGCAGCAAGUACGAGUCCGAUGGUGUUGCCCGCUCUGAGGAGCUGGAGGAGGCCAA ((((..((((((.(((....(((..((((((((..((..(((....)))..))....))).)))))..)))....((....))..))).))))))((((....)))).......)))).. ( -43.90) >consensus GGCUGAGGGCAAGGCUGAUCUGCAGCGUCAACUGAGCAAGGCCAACGCCGAGGCCCAGGUCUGGCGCAGCAAGUACGAGUCCGAUGGUGUUGCCCGCUCUGAGGAGCUGGAGGAGGCCAA ((((..((((((.((((...(((.((((((.(((.((..(((....)))...)).)))...)))))).)))....((....)).)))).))))))((((....)))).......)))).. (-46.21 = -45.97 + -0.24)


| Location | 16,781,037 – 16,781,157 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -50.50 |
| Consensus MFE | -42.30 |
| Energy contribution | -42.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16781037 120 - 22407834 AGCUGCAGAUCCUCCACCUCGGUGGACAGACGCUGCUUGGUCUUCUCCAGGCCAAUGCACUUCUGGUUGAGGGACUCGAUGGUCUCCUCGGCCUCGGCCAAACGGGCCUGCAGCUUCCUC ((((((((..(((((((....)))))((((.(.((((((((((.....))))))).)))).))))(((((((((((....)))).))))(((....))).)))))..))))))))..... ( -58.90) >DroVir_CAF1 1315 120 - 1 AGCUGCAAGUCCUCGACUUCGGUGGACAGGCGCUGCUUGGUCUUCUCCAGGCCGAUGCACUUCUGGUUGAGCGACUCGAUGGUCUCCUCAGCCUCGGCAAGGCGGGCCUGCAGCUUCCUC ((((((..(((((((....))).))))((((.(((((((((((.....)))))..(((......(((((((.((((....))))..)))))))...)))))))))))))))))))..... ( -54.40) >DroPse_CAF1 4135 120 - 1 AGCUGGAGAUCCUCGACUUCGGUGGACAGACGCUGCUUGGUCUUCUCCAGGCCAAUGCACUUCUGGUUAAGGGACUCGAUGGUCUCCUCAGCCUCGGCGAGACGGGCCUGCAGCUUGCGC (((((.((..((((.((....)).))....(((((((((((((.....))))))).))......((((.(((((((....)))).))).))))..))))....))..)).)))))..... ( -46.70) >DroWil_CAF1 731 120 - 1 AAUUGCAGAUCCUCCACUUCAGUGGACAGACGCUGCUUGGUCUUCUCCAGACCAAUGCACUUCUGGUUGAGGGACUCAAUGGUCUCUUCAGCCUCAGCCAAACGGGCCUGCAACUUCCUC ..((((((..(((((((....))))).....((((((((((((.....))))))).))......((((((((((((....)))).))))))))..))).....))..))))))....... ( -47.70) >DroMoj_CAF1 1298 120 - 1 AGCUGCAAGUCCUCCACUUCGGUGGACAGGCGCUGCUUGGUCUUCUCCAGGCCGAUGCACUUCUGGUUGAGCGACUCAAUGGUCUCCUCAGCCUCGGCAAGGCGGGCCUGCAACUUCCUC ((.((((.(((((((((....))))).....(((((((((......))))))...(((......(((((((.((((....))))..)))))))...))).))))))).)))).))..... ( -48.60) >DroPer_CAF1 4007 120 - 1 AGCUGGAGAUCCUCGACUUCGGUGGACAGACGCUGCUUGGUCUUCUCCAGGCCAAUGCACUUCUGGUUAAGGGACUCGAUGGUCUCCUCAGCCUCGGCGAGACGGGCCUGCAGCUUGCGC (((((.((..((((.((....)).))....(((((((((((((.....))))))).))......((((.(((((((....)))).))).))))..))))....))..)).)))))..... ( -46.70) >consensus AGCUGCAGAUCCUCCACUUCGGUGGACAGACGCUGCUUGGUCUUCUCCAGGCCAAUGCACUUCUGGUUGAGGGACUCGAUGGUCUCCUCAGCCUCGGCAAGACGGGCCUGCAGCUUCCUC ((((((((.(((((.((....)).)).....((((((((((((.....))))))).))......((((((((((((....)))).))))))))..))).....))).))))))))..... (-42.30 = -42.97 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:37 2006