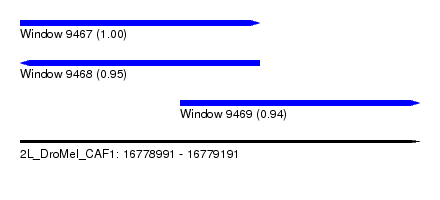
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,778,991 – 16,779,191 |
| Length | 200 |
| Max. P | 0.998990 |

| Location | 16,778,991 – 16,779,111 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -39.52 |
| Energy contribution | -40.05 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
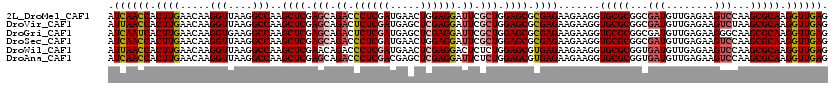
>2L_DroMel_CAF1 16778991 120 + 22407834 AUCAACCACUUGAACAAGGUUAAGGCCAAGCUCGAGCAGACCCUCGAUGAACUGGAGGAUUCGCUGGAGCGCGAGAAGAAGGUGCGCGGCGAUGUUGAGAAGUCCAAGCGCAAGGUUGAG .((((((.((((.....(((....)))..((((.(((..(.((((.(.....).)))).)..))).)))).)))).......(((((...(((........)))...))))).)))))). ( -43.60) >DroVir_CAF1 9113 120 + 1 AUUAACCACUUGAACAAGGUUAAGGCCAAGCUCGAGCAGACUCUCGAUGAGCUCGAGGAUUCGCUGGAGCGCGAGAAGAAGGUGCGCGGCGAUGUUGAGAAGUCUAAGCGCAAGGUUGAG .((((((.((((.....(((....)))..((((.(((..(..(((((.....)))))..)..))).)))).)))).......(((((...(((........)))...))))).)))))). ( -43.90) >DroGri_CAF1 9311 120 + 1 AUCAAUCACUUGAACAAGGUGAAGGCCAAGCUCGAGCAGACUCUCGAUGAGCUCGAGGAUUCGCUGGAGCGCGAGAAGAAGGUGCGCGGCGAUGUUGAGAAGGGCAAGCGCAAGGUUGAG .((((((.(((....)))(((...(((...(((.((((...((((((.....))))))..((((((..((((.........)))).)))))))))))))...)))...)))..)))))). ( -42.90) >DroSec_CAF1 8598 120 + 1 AUCAACCACUUGAACAAGGUUAAGGCCAAGCUCGAGCAGACCCUCGAUGAACUGGAGGAUUCGCUGGAGCGCGAGAAGAAGGUGCGCGGCGAUGUUGAGAAGUCCAAGCGCAAGGUUGAG .((((((.((((.....(((....)))..((((.(((..(.((((.(.....).)))).)..))).)))).)))).......(((((...(((........)))...))))).)))))). ( -43.60) >DroWil_CAF1 8712 120 + 1 AUUAACCACUUGAACAAGGUUAAGGCCAAGCUCGAACAGACCCUCGAUGAACUCGAGGACUCUCUGGAGCGUGAGAAGAAGGUGCGCGGUGAUGUUGAGAAGUCCAAGCGCAAGGUUGAG .((((((.(((..((..(((....)))..((((...((((.((((((.....))))))....))))))))))...)))....(((((...(((........)))...))))).)))))). ( -37.30) >DroAna_CAF1 8823 120 + 1 AUCAACCACUUGAACAAGGUUAAGGCCAAGCUCGAGCAGACCCUCGACGAGCUCGAGGAUUCUCUGGAGCGUGAGAAGAAGGUGCGCGGUGAUGUUGAGAAGUCCAAGCGCAAGGUUGAG .((((((.(((..((..(((....)))..((((.((.(((.((((((.....))))))..))))).))))))...)))....(((((...(((........)))...))))).)))))). ( -43.20) >consensus AUCAACCACUUGAACAAGGUUAAGGCCAAGCUCGAGCAGACCCUCGAUGAACUCGAGGAUUCGCUGGAGCGCGAGAAGAAGGUGCGCGGCGAUGUUGAGAAGUCCAAGCGCAAGGUUGAG .((((((.((((.....(((....)))..((((.(((.((.((((((.....)))))).)).))).)))).)))).......(((((...(((........)))...))))).)))))). (-39.52 = -40.05 + 0.53)
| Location | 16,778,991 – 16,779,111 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -30.79 |
| Energy contribution | -33.18 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16778991 120 - 22407834 CUCAACCUUGCGCUUGGACUUCUCAACAUCGCCGCGCACCUUCUUCUCGCGCUCCAGCGAAUCCUCCAGUUCAUCGAGGGUCUGCUCGAGCUUGGCCUUAACCUUGUUCAAGUGGUUGAU .((((((....((((((((...........((((((...........)))((((.((((.((((((.........)))))).)))).))))..))).........)))))))))))))). ( -40.25) >DroVir_CAF1 9113 120 - 1 CUCAACCUUGCGCUUAGACUUCUCAACAUCGCCGCGCACCUUCUUCUCGCGCUCCAGCGAAUCCUCGAGCUCAUCGAGAGUCUGCUCGAGCUUGGCCUUAACCUUGUUCAAGUGGUUAAU ...((((........(((((........((((.((((...........))))....))))...(((((.....))))))))))(((.((((..((......))..)))).)))))))... ( -32.70) >DroGri_CAF1 9311 120 - 1 CUCAACCUUGCGCUUGCCCUUCUCAACAUCGCCGCGCACCUUCUUCUCGCGCUCCAGCGAAUCCUCGAGCUCAUCGAGAGUCUGCUCGAGCUUGGCCUUCACCUUGUUCAAGUGAUUGAU .((((...(((((..((.............)).)))))........((((((((.((((.(..(((((.....)))))..).)))).))))(((..(........)..))))))))))). ( -32.12) >DroSec_CAF1 8598 120 - 1 CUCAACCUUGCGCUUGGACUUCUCAACAUCGCCGCGCACCUUCUUCUCGCGCUCCAGCGAAUCCUCCAGUUCAUCGAGGGUCUGCUCGAGCUUGGCCUUAACCUUGUUCAAGUGGUUGAU .((((((....((((((((...........((((((...........)))((((.((((.((((((.........)))))).)))).))))..))).........)))))))))))))). ( -40.25) >DroWil_CAF1 8712 120 - 1 CUCAACCUUGCGCUUGGACUUCUCAACAUCACCGCGCACCUUCUUCUCACGCUCCAGAGAGUCCUCGAGUUCAUCGAGGGUCUGUUCGAGCUUGGCCUUAACCUUGUUCAAGUGGUUAAU ..........(((((((((...............................((((.(.(((.(((((((.....)))))))))).)..))))..((......))..)))))))))...... ( -31.10) >DroAna_CAF1 8823 120 - 1 CUCAACCUUGCGCUUGGACUUCUCAACAUCACCGCGCACCUUCUUCUCACGCUCCAGAGAAUCCUCGAGCUCGUCGAGGGUCUGCUCGAGCUUGGCCUUAACCUUGUUCAAGUGGUUGAU .((((((....((((((((...............................((((.(((((.(((((((.....)))))))))).)).))))..((......))..)))))))))))))). ( -36.20) >consensus CUCAACCUUGCGCUUGGACUUCUCAACAUCGCCGCGCACCUUCUUCUCGCGCUCCAGCGAAUCCUCGAGCUCAUCGAGGGUCUGCUCGAGCUUGGCCUUAACCUUGUUCAAGUGGUUGAU .((((((....((((((((...........((((((...........)))((((.((((.((((((((.....)))))))).)))).))))..))).........)))))))))))))). (-30.79 = -33.18 + 2.39)

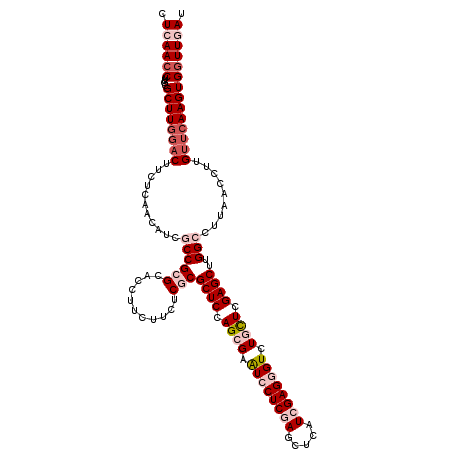
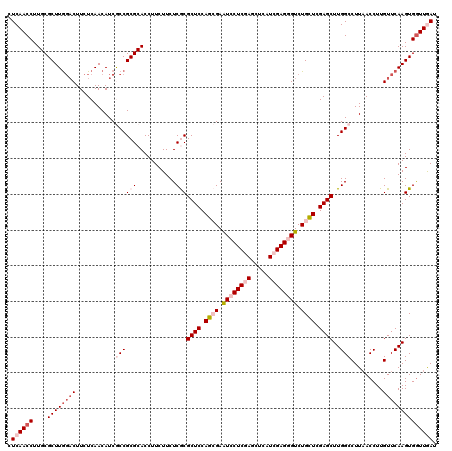
| Location | 16,779,071 – 16,779,191 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -40.24 |
| Energy contribution | -41.18 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16779071 120 + 22407834 GGUGCGCGGCGAUGUUGAGAAGUCCAAGCGCAAGGUUGAGGGCGACCUCAAGCUCACCCAGGAGGCUGUUGCCGAUCUGGAGCGCAACAAGAAGGAGCUCGAGCAGACCAUCCAGCGCAA ..(((((.(.(((((((((...(((..((((.((.((((((....)))))).))...(((((.(((....)))..))))).))))........))).)))))......))))).))))). ( -47.00) >DroVir_CAF1 9193 120 + 1 GGUGCGCGGCGAUGUUGAGAAGUCUAAGCGCAAGGUUGAGGGUGACCUCAAGCUGACCCAGGAGGCUGUUUCCGAUCUGGAGCGCAACAAGAAGGAGUUGGAGCAGACCAUCCAGCGUAA (((((((...(((........)))...)))).((.((((((....)))))).)).)))..((((((((((.(((((((....(.......)..))).))))))))).)).)))....... ( -42.80) >DroGri_CAF1 9391 120 + 1 GGUGCGCGGCGAUGUUGAGAAGGGCAAGCGCAAGGUUGAGGGUGACCUCAAGCUGACCCAGGAGGCUGUUGCCGAUCUGGAGCGCAACAAGAAGGAGCUGGAGCAGACCAUUCAGCGCAA ..((((((((..((((......)))).((((.((.((((((....)))))).))...(((((.(((....)))..))))).))))...........)))((......)).....))))). ( -48.00) >DroSec_CAF1 8678 120 + 1 GGUGCGCGGCGAUGUUGAGAAGUCCAAGCGCAAGGUUGAGGGCGACCUCAAGCUCACCCAGGAGGCUGUUGCCGAUCUGGAGCGCAACAAGAAGGAGCUCGAGCAGACCAUCCAGCGCAA ..(((((.(.(((((((((...(((..((((.((.((((((....)))))).))...(((((.(((....)))..))))).))))........))).)))))......))))).))))). ( -47.00) >DroWil_CAF1 8792 120 + 1 GGUGCGCGGUGAUGUUGAGAAGUCCAAGCGCAAGGUUGAGGGUGACCUUAAGUUGACUCAGGAGGCUGUCGCUGAUCUUGAGCGCAACAAGAAGGAGUUGGAACAGACCAUCCAACGCAA ..(((((...(((........)))...)))))...((((((....))))))((((.((((((((((....)))..)))))))..))))........((((((........)))))).... ( -41.00) >DroAna_CAF1 8903 120 + 1 GGUGCGCGGUGAUGUUGAGAAGUCCAAGCGCAAGGUUGAGGGAGACCUCAAGCUCACCCAGGAGGCUGUUGCCGAUCUGGAGCGCAACAAGAAGGAAUUGGAGCAGACCAUCCAGCGCAA ..(((((.(.(((((((.....(((((((((.((.((((((....)))))).))...(((((.(((....)))..))))).))))..(.....)...))))).)))..))))).))))). ( -48.40) >consensus GGUGCGCGGCGAUGUUGAGAAGUCCAAGCGCAAGGUUGAGGGUGACCUCAAGCUCACCCAGGAGGCUGUUGCCGAUCUGGAGCGCAACAAGAAGGAGCUGGAGCAGACCAUCCAGCGCAA ..(((((.(.((((.............((((.((.((((((....)))))).))...(((((.(((....)))..))))).))))...........((....))....))))).))))). (-40.24 = -41.18 + 0.95)
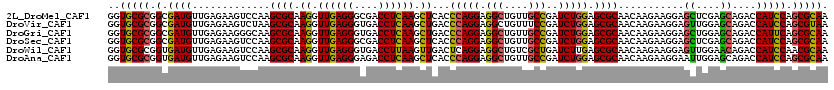
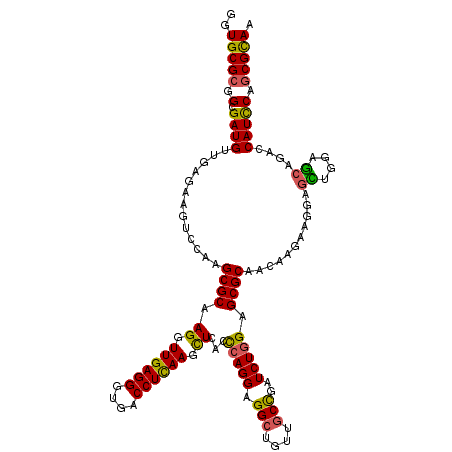
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:32 2006