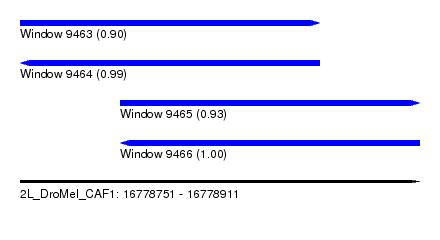
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,778,751 – 16,778,911 |
| Length | 160 |
| Max. P | 0.997812 |

| Location | 16,778,751 – 16,778,871 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -33.74 |
| Energy contribution | -34.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16778751 120 + 22407834 UUCCAGCAGAAGAAGAAGGCCGAUCAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGC .....((..........((((((((....))))..((((((...((......)).....((((((........))))))))))))..........)))).....(((((....))))))) ( -37.10) >DroVir_CAF1 8873 120 + 1 UUCCAGCAGAAGAAGAAGGCCGAUCAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAACAGGAUAAGGCCACCAAGGAUCACCAGAUCCGC .....((..........((((((((....))))..((((((......((((.((((..........))))..))))...))))))..........)))).....(((((....))))))) ( -37.10) >DroGri_CAF1 9071 120 + 1 UUCCAGCAGAAGAAGAAGGCCGAUCAAGAGAUCUCUGGCCUGAAGAAGGACAUCGAAGAUAUGGAGCUGAACAUCCAGAAGGCCGAACAGGAUAAGGCCACCAAGGAUCACCAGAUCCGC .....((..........((((((((....))))..((((((...((......)).......((((........))))..))))))..........)))).....(((((....))))))) ( -33.40) >DroWil_CAF1 8472 120 + 1 UUCCAACAGAAGAAGAAGGCCGACCAGGAGAUCUCUGGCUUGAAGAAGGAUAUCGAAGAUCUGGAAUUGAACAUCCAGAAGGCCGAGCAAGACAAGGCCACCAAGGAUCACCAGAUCCGC .................((((..(((((.....)))))((((..........(((....((((((........))))))....))).))))....)))).....(((((....))))).. ( -32.10) >DroMoj_CAF1 9068 120 + 1 UUCCAGCAGAAGAAGAAGGCCGAUCAGGAGAUCUCCGGCUUGAAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAGCAGGAUAAGGCCACCAAGGAUCACCAGAUCCGC .....((..........((((((((....))))(((.(((((..((......)).....((((((........))))))....))))).)))...)))).....(((((....))))))) ( -37.80) >DroAna_CAF1 8583 120 + 1 UUCCAGCAGAAGAAGAAGGCCGACCAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAGCUGAAUGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGC .....((..........((((..(((((.....)))))((((..((......))..((.((((((.(.....)))))))...))...))))....)))).....(((((....))))))) ( -36.70) >consensus UUCCAGCAGAAGAAGAAGGCCGAUCAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGC .....((..........((((((((....))))..((((((...((......)).....((((((........))))))))))))..........)))).....(((((....))))))) (-33.74 = -34.05 + 0.31)


| Location | 16,778,751 – 16,778,871 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -36.68 |
| Energy contribution | -36.35 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16778751 120 - 22407834 GCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUGAUCGGCCUUCUUCUUCUGCUGGAA ((.((((.((.((((.(((((((((..(.....)..)))).((((((........))))))....................))))).)))).)).)))).)).....(((......))). ( -38.90) >DroVir_CAF1 8873 120 - 1 GCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGUUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUGAUCGGCCUUCUUCUUCUGCUGGAA (((((((....)))))..((.((((...........(((((((((((........)))))).....((......))...)))))...((((....)))))))).)).......))..... ( -37.70) >DroGri_CAF1 9071 120 - 1 GCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGUUCGGCCUUCUGGAUGUUCAGCUCCAUAUCUUCGAUGUCCUUCUUCAGGCCAGAGAUCUCUUGAUCGGCCUUCUUCUUCUGCUGGAA (((((((....)))))..((.((((...........(((((...(((((((.((........))..)))))))......)))))...((((....)))))))).)).......))..... ( -34.80) >DroWil_CAF1 8472 120 - 1 GCGGAUCUGGUGAUCCUUGGUGGCCUUGUCUUGCUCGGCCUUCUGGAUGUUCAAUUCCAGAUCUUCGAUAUCCUUCUUCAAGCCAGAGAUCUCCUGGUCGGCCUUCUUCUUCUGUUGGAA ((.((((.((.((((.(((((((((..(.....)..)))).((((((........))))))....................))))).)))).)).)))).)).....(((......))). ( -39.10) >DroMoj_CAF1 9068 120 - 1 GCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGCUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAAGCCGGAGAUCUCCUGAUCGGCCUUCUUCUUCUGCUGGAA (((((((....)))))..((.((((...(((.((((((...((((((........))))))...))))((........)).)).)))((((....)))))))).)).......))..... ( -36.90) >DroAna_CAF1 8583 120 - 1 GCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACAUUCAGCUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUGGUCGGCCUUCUUCUUCUGCUGGAA ((((((..(((.((.....)).)))..)))).))..((((....(((((((.((.........)).))))))).......((((((.(....)))))))))))....(((......))). ( -39.60) >consensus GCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUGAUCGGCCUUCUUCUUCUGCUGGAA ((.((((.((.((((..((((((((...........)))).((((((........))))))....................))))..)))).)).)))).)).....(((......))). (-36.68 = -36.35 + -0.33)


| Location | 16,778,791 – 16,778,911 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -32.80 |
| Energy contribution | -32.97 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
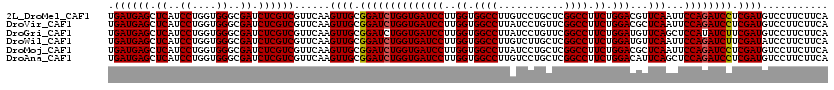
>2L_DroMel_CAF1 16778791 120 + 22407834 UGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACCAGGAUGAGCUCAUCA ............(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((....)).))).))).)). ( -35.30) >DroVir_CAF1 8913 120 + 1 UGAAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAACAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACCAGGAUGAGCUCAUCA ............(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((....)).))).))).)). ( -35.30) >DroGri_CAF1 9111 120 + 1 UGAAGAAGGACAUCGAAGAUAUGGAGCUGAACAUCCAGAAGGCCGAACAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACCAGGAUGAGCUCAUCA .......................(((((...(((((....((((...........)))).....(((((....)))))((..((((.....))))...))......)))))))))).... ( -30.80) >DroWil_CAF1 8512 120 + 1 UGAAGAAGGAUAUCGAAGAUCUGGAAUUGAACAUCCAGAAGGCCGAGCAAGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACCAGGAUGAGCUCAUCA (.(((..((..........((((((........)))))).((((...........)))).))..(((((....)))))....))).)..((.(((.(((.((....)).))).))).)). ( -33.60) >DroMoj_CAF1 9108 120 + 1 UGAAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAGCAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACCAGGAUGAGCUCAUCA ............(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((....)).))).))).)). ( -35.30) >DroAna_CAF1 8623 120 + 1 UGAAGAAGGACAUCGAGGAUCUGGAGCUGAAUGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACCAGGAUGAGCUCAUCA ............(((((..((((((.(.....))))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((....)).))).))).)). ( -35.40) >consensus UGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACCAGGAUGAGCUCAUCA (.(((..((..........((((((........)))))).((((...........)))).))..(((((....)))))....))).)..((.(((.(((.((....)).))).))).)). (-32.80 = -32.97 + 0.17)
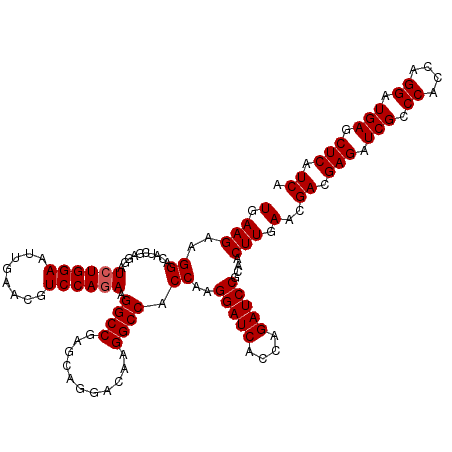

| Location | 16,778,791 – 16,778,911 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -39.14 |
| Energy contribution | -39.08 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16778791 120 - 22407834 UGAUGAGCUCAUCCUGGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCA .((((((.((..((....))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))........... ( -41.90) >DroVir_CAF1 8913 120 - 1 UGAUGAGCUCAUCCUGGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGUUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUUCA .((((((.((..((....))..)).))))))......((((.((((((((((((((..((.((((...........)))).)).)))...)))...)))))))).))))........... ( -40.60) >DroGri_CAF1 9111 120 - 1 UGAUGAGCUCAUCCUGGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGUUCGGCCUUCUGGAUGUUCAGCUCCAUAUCUUCGAUGUCCUUCUUCA .((((((.((..((....))..)).))))))(((.(((.((.(((.((((..((((..((.((((...........)))).)).))))..)))).))).)).))).)))........... ( -35.50) >DroWil_CAF1 8512 120 - 1 UGAUGAGCUCAUCCUGGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCUUGCUCGGCCUUCUGGAUGUUCAAUUCCAGAUCUUCGAUAUCCUUCUUCA .((((((.((..((....))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))........... ( -39.60) >DroMoj_CAF1 9108 120 - 1 UGAUGAGCUCAUCCUGGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGCUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUUCA .((((((.((..((....))..)).))))))......((((.((((((((((((((..((.((((...........)))).)).)))...)))...)))))))).))))........... ( -40.60) >DroAna_CAF1 8623 120 - 1 UGAUGAGCUCAUCCUGGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACAUUCAGCUCCAGAUCCUCGAUGUCCUUCUUCA .((((((.((..((....))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))........... ( -41.90) >consensus UGAUGAGCUCAUCCUGGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCA .((((((.((..((....))..)).))))))......((((.((((((((((((((..((.((((...........)))).)).)))...)))...)))))))).))))........... (-39.14 = -39.08 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:29 2006