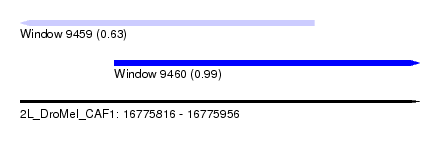
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,775,816 – 16,775,956 |
| Length | 140 |
| Max. P | 0.994090 |

| Location | 16,775,816 – 16,775,919 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 72.07 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -6.32 |
| Energy contribution | -8.90 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16775816 103 - 22407834 CUCAAUCAAACCCUGACGAAUU--GUGAAAUUCGAUGGUGCUUGAGU-CAAGUUAAGCGGACUGGGAAACAUUGUCCAGAUUCAAACAAUAGAGUGCAGAUGAUGU .(((.((..((.(((.((((((--....))))))....((.((((((-(..(.....)((((..(....)...)))).))))))).)).))).))...)))))... ( -24.90) >DroSec_CAF1 5448 102 - 1 CUCAAUCAAACCCUGACGAAUU--GUGAAAUUCGAUGGUGCUUGAGU-CAAGUUAAGCGGACUGGGAAACAUUGUCCAGAUUCAAGCA-UAAAGUGCAGAUGAUGU ....((((....(((.((((((--....))))))...((((((((((-(..(.....)((((..(....)...)))).))))))))))-)......))).)))).. ( -32.50) >DroSim_CAF1 5981 104 - 1 NNNNNNNNNNNNNNNNNNNNNN--NNNNNNNNNNNNNNNNNNNGAUUCCAAGUUAAGCGGACUGGGAAACAUUUUCCAGAUUCAAGCAGUAAAGUGCAGAUGAUGU ......................--...........................((((...((((((((((....))))))).)))..(((......)))...)))).. ( -11.50) >DroEre_CAF1 5462 102 - 1 CUCAAUCAAGCCCU-ACGAACU--ACGAAAUUCGAUAGUGCUUGAGU-CAGGUCCAGCAGAUUGGGAAACAUUCUGCGGACUGAAGCAGUAGAGUGCAGAUGAUGU .....((((((.((-(((((..--......)))).))).))))))((-(((((((.(((((...(....)..))))))))))...(((......)))...)))).. ( -35.30) >DroYak_CAF1 5411 105 - 1 CUCAAUCAAACCCUGACGAAUUCGAUGAAAUUCGAUAGGGCUUGAGU-CGAGUUAAGCAGUCAGGAAGACAUUGUCGAGAUUGAAGCAGUAGAGUGCAGAUGAUGU .((((((....((((((....((((......))))....((((((..-....)))))).))))))..(((...)))..)))))).(((......)))......... ( -28.80) >consensus CUCAAUCAAACCCUGACGAAUU__AUGAAAUUCGAUAGUGCUUGAGU_CAAGUUAAGCGGACUGGGAAACAUUGUCCAGAUUCAAGCAGUAGAGUGCAGAUGAUGU ..........(((...((((((......)))))).....((((((.......)))))).....)))..((((((((...(((..........)))...)))))))) ( -6.32 = -8.90 + 2.58)

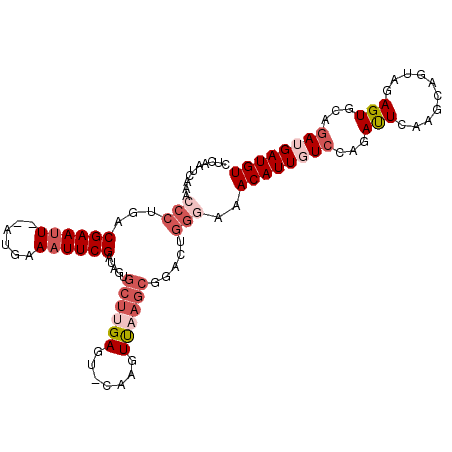
| Location | 16,775,849 – 16,775,956 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -27.02 |
| Energy contribution | -26.15 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16775849 107 + 22407834 AAUGUUUCCCAGUCCGCUUAACUUGACUCAAGCACCAUCGAAUUUCAC--AAUUCGUCAGGGUUUGAUUGAGGUUUAGGCCAAUCGGUAGAUAUUAUCAAGACUCACUU (((((((.((.((..(((((((((.(.((((((.((..((((((....--))))))...)))))))).).))).))))))..)).)).))))))).............. ( -27.20) >DroSec_CAF1 5480 107 + 1 AAUGUUUCCCAGUCCGCUUAACUUGACUCAAGCACCAUCGAAUUUCAC--AAUUCGUCAGGGUUUGAUUGAGGUUUAGGCCAAUCGGUAGAUAUUAUCAAGACUCACUU (((((((.((.((..(((((((((.(.((((((.((..((((((....--))))))...)))))))).).))).))))))..)).)).))))))).............. ( -27.20) >DroEre_CAF1 5495 103 + 1 AAUGUUUCCCAAUCUGCUGGACCUGACUCAAGCACUAUCGAAUUUCGU--AGUUCGU-AGGGCUUGAUUGAGGUUUUGGCCAAU-GGUAGAUAUUGUCAAGACU--CUG (((((((.(((....((..(((((.(.((((((.(((.((((((....--)))))))-)).)))))).).))).))..))...)-)).))))))).........--... ( -33.30) >DroYak_CAF1 5444 106 + 1 AAUGUCUUCCUGACUGCUUAACUCGACUCAAGCCCUAUCGAAUUUCAUCGAAUUCGUCAGGGUUUGAUUGAGGUUUUGGCCAAUCGGUAGAUAUUAUCUAGAC---CUC (((((((..((((..(((...(((((.(((((((((..(((((((....)))))))..)))))))))))))).....)))...)))).)))))))........---... ( -36.90) >consensus AAUGUUUCCCAGUCCGCUUAACUUGACUCAAGCACCAUCGAAUUUCAC__AAUUCGUCAGGGUUUGAUUGAGGUUUAGGCCAAUCGGUAGAUAUUAUCAAGACU__CUU (((((((.((.((..(((((((((.(.((((((.((..((((((......))))))...)))))))).).))).))))))..)).)).))))))).............. (-27.02 = -26.15 + -0.87)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:23 2006