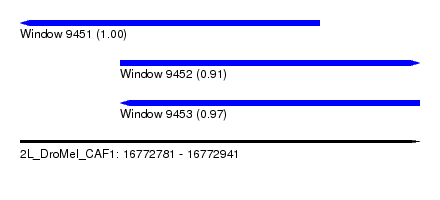
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,772,781 – 16,772,941 |
| Length | 160 |
| Max. P | 0.998076 |

| Location | 16,772,781 – 16,772,901 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -46.28 |
| Consensus MFE | -42.35 |
| Energy contribution | -42.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
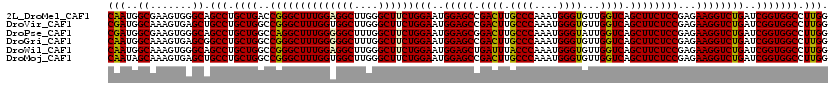
>2L_DroMel_CAF1 16772781 120 - 22407834 CUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGGGGAACAUAGACUCUUCCUCCAGGAUGGACAAGAUACCCAU ..((((((((((((..(((((.((((.((((....))))...)))).)))))))).)))))((((..((.((..(((.((((((.........)))))).))))).))..)))).)))). ( -46.30) >DroVir_CAF1 2617 120 - 1 CUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGGGGAACAUAGACUCUUCCUCAAGAAUCGACAAGAUACCCAU ..((((((((((((..(((((.((((.((((....))))...)))).)))))))).)))))((((..(((((...(((((((((.........))))))))).)))))..)))).)))). ( -51.00) >DroPse_CAF1 2613 120 - 1 CUUUGGCUUCUGGAAUGGAGCGGACUUGCCCAAAUGGGUAUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGGGGAACAUAGACUCUUCCUCCAGGAUGGACAGGAUACCCAU ...(((((((((((..(((((.(((((((((....)))))..)))).)))))))).)))))((((..((.((..(((.((((((.........)))))).))))).))..))))..))). ( -43.40) >DroGri_CAF1 2573 120 - 1 CUUUGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGGGGAACAUAGACUCUUCCUCCAGGAUGGACAAGAUACCCAU ..((((((((......)))))))).........(((((((((.(((.(((...((((..(....)..)))).)))(((((((((.........)))).)))))...)))..))))))))) ( -42.90) >DroWil_CAF1 2392 120 - 1 CUUGGGCUUCUGGAAUGGAGCUGAUUUACCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGGGGAACAUAGACUCUUCUUCCAGGAUGGACAAGAUACCCAU ((((..((((((((..(((((((((((((((....)))))..))))))))))....((((((((((....((..((....)).)).))))).)))))))))))).)..))))........ ( -46.50) >DroAna_CAF1 2435 120 - 1 CUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGGGGAACAUAGACUCUUCUUCAAGGAUGGACAGGAUACCCAU ..((((((((((((..(((((.((((.((((....))))...)))).)))))))).)))))((((..((.((..((((((((((.........)))))))))))).))..)))).)))). ( -47.60) >consensus CUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGGGGAACAUAGACUCUUCCUCCAGGAUGGACAAGAUACCCAU ..((((((((((((..(((((.((((.((((....))))...)))).)))))))).)))))((((..((.((..(((.((((((.........)))))).))))).))..)))).)))). (-42.35 = -42.13 + -0.22)

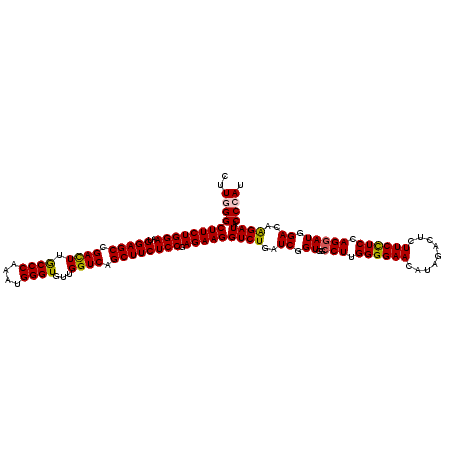
| Location | 16,772,821 – 16,772,941 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -32.37 |
| Energy contribution | -32.32 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16772821 120 + 22407834 CCAAGGCCACCGAUCAGACCUUCUCGGAGAAGCUGACCAACACCCAUUUGGGCAAGUCGGCUCCAUUCCAGAAGCCCAAGCCUCCAAAGCCCGGUCAGCAGGCUGCCCACUUCGCCAUUG ....((((.((((..........))))....(((((((.........((((((...((((.......)).)).))))))((.......))..))))))).))))................ ( -34.50) >DroVir_CAF1 2657 120 + 1 CCAAGGCCACCGAUCAGACCUUCUCGGAGAAGCUGACCAACACCCAUUUGGGCAAGUCGGCUCCAUUCCAGAAGCCCAAGCCACCAAAGCCCGGCCAGCAGGCAGCUCACUUUGCCAUCG ....((((........(..(((((.((((.(((((((.....(((....)))...)))))))...)))))))))..)..((.......))..))))....(((((......))))).... ( -37.00) >DroPse_CAF1 2653 120 + 1 CCAAGGCCACCGAUCAGACCUUCUCGGAGAAGCUGACCAAUACCCAUUUGGGCAAGUCCGCUCCAUUCCAGAAGCCAAAGCCCCCAAAGCCUGGCCAGCAGGCUGCCCACUUCGCCAUCG ....(((............(((((.((((.(((.(((.....(((....)))...))).)))...))))))))).............((((((.....)))))).........))).... ( -32.30) >DroGri_CAF1 2613 120 + 1 CCAAGGCCACCGAUCAGACCUUCUCGGAGAAGCUGACCAACACCCAUUUGGGCAAGUCGGCUCCAUUCCAGAAGCCAAAGCCCCCAAAGCCCGGCCAGCAGGCCGCUCACUUUGCCAUUG ....(((............(((((.((((.(((((((.....(((....)))...)))))))...)))))))))...........((((..(((((....)))))....))))))).... ( -36.30) >DroWil_CAF1 2432 120 + 1 CCAAGGCCACCGAUCAGACCUUCUCGGAGAAGCUGACCAACACCCAUUUGGGUAAAUCAGCUCCAUUCCAGAAGCCCAAGCCUCCAAAGCCCGGCCAGCAGGCUGCCCACUUUGCCAUUG ....((((........(..(((((.((((.((((((.....((((....))))...))))))...)))))))))..)..((.......))..))))....(((..........))).... ( -34.30) >DroMoj_CAF1 2678 120 + 1 CCAAGGCCACCGAUCAGACCUUCUCGGAGAAGCUGACCAACACCCAUUUGGGCAAGUCGGCUCCAUUCCAGAAGCCCAAGCCACCAAAGCCCGGCCAGCAGGCAGCUCACUUUGCUAUUG ....((((........(..(((((.((((.(((((((.....(((....)))...)))))))...)))))))))..)..((.......))..))))((((((........)))))).... ( -34.70) >consensus CCAAGGCCACCGAUCAGACCUUCUCGGAGAAGCUGACCAACACCCAUUUGGGCAAGUCGGCUCCAUUCCAGAAGCCCAAGCCACCAAAGCCCGGCCAGCAGGCUGCCCACUUUGCCAUUG ....((((...........(((((.((((.(((((((.....(((....)))...)))))))...))))))))).....((.......))..))))....(((..........))).... (-32.37 = -32.32 + -0.05)

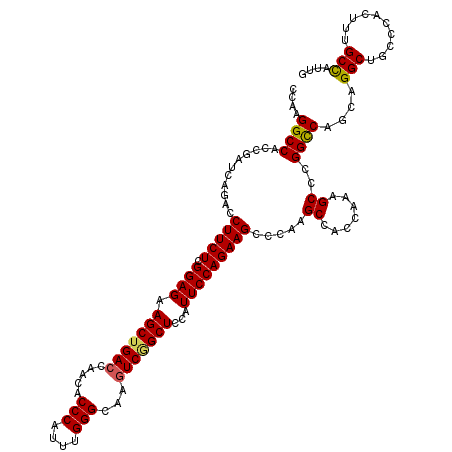
| Location | 16,772,821 – 16,772,941 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -49.63 |
| Consensus MFE | -46.23 |
| Energy contribution | -45.82 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
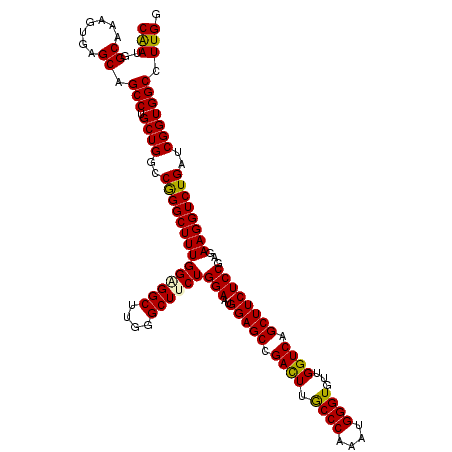
>2L_DroMel_CAF1 16772821 120 - 22407834 CAAUGGCGAAGUGGGCAGCCUGCUGACCGGGCUUUGGAGGCUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGG (((.(((.((((.(((.(((((.....)))))((..(((((....)))))..)).....))).))))((((....)))).((((((((((((.....))))..))))))))..)))))). ( -49.50) >DroVir_CAF1 2657 120 - 1 CGAUGGCAAAGUGAGCUGCCUGCUGGCCGGGCUUUGGUGGCUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGG (((.(((.....((((..((.(((.....)))...))..))))((.((((((((..(((((.((((.((((....))))...)))).)))))))).))))).)).........)))))). ( -48.30) >DroPse_CAF1 2653 120 - 1 CGAUGGCGAAGUGGGCAGCCUGCUGGCCAGGCUUUGGGGGCUUUGGCUUCUGGAAUGGAGCGGACUUGCCCAAAUGGGUAUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGG (((.(((.....((....)).((((..((((((((((((((....))))))(((..(((((.(((((((((....)))))..)))).))))))))...))))))))..)))).)))))). ( -49.60) >DroGri_CAF1 2613 120 - 1 CAAUGGCAAAGUGAGCGGCCUGCUGGCCGGGCUUUGGGGGCUUUGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGG .....((.......))((((.((((..((((((((((((((....))))))(((..(((((.((((.((((....))))...)))).))))))))...))))))))..)))))))).... ( -51.30) >DroWil_CAF1 2432 120 - 1 CAAUGGCAAAGUGGGCAGCCUGCUGGCCGGGCUUUGGAGGCUUGGGCUUCUGGAAUGGAGCUGAUUUACCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGG (((.(((.....((....)).((((..((((((((((((((....))))))(((..(((((((((((((((....)))))..)))))))))))))...))))))))..)))).)))))). ( -52.10) >DroMoj_CAF1 2678 120 - 1 CAAUAGCAAAGUGAGCUGCCUGCUGGCCGGGCUUUGGUGGCUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGG ....(((.......)))(((.((((..((((((((((.((((..((((((......))))))((((.((((....))))...))))))))...))))...))))))..)))))))..... ( -47.00) >consensus CAAUGGCAAAGUGAGCAGCCUGCUGGCCGGGCUUUGGAGGCUUGGGCUUCUGGAAUGGAGCCGACUUGCCCAAAUGGGUGUUGGUCAGCUUCUCCGAGAAGGUCUGAUCGGUGGCCUUGG (((..((.......)).(((.((((..((((((((((((((....))))))(((..(((((.((((.((((....))))...)))).))))))))...))))))))..))))))).))). (-46.23 = -45.82 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:16 2006