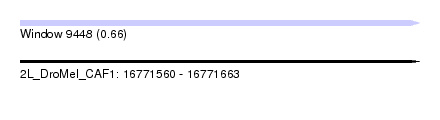
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,771,560 – 16,771,663 |
| Length | 103 |
| Max. P | 0.660578 |

| Location | 16,771,560 – 16,771,663 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -15.12 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
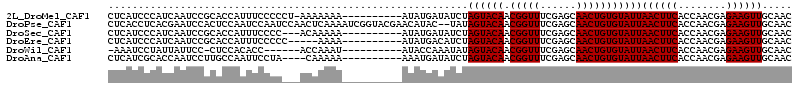
>2L_DroMel_CAF1 16771560 103 + 22407834 CUCAUCCCAUCAAUCCGCACCAUUUCCCCCU-AAAAAAA----------AUAUGAUAUCUAGUACAACGGUUUCGAGCAACUGUGUAUUAACUUCACCAACGAGAAGUUGCAAC ................(((............-.......----------..........(((((((.(((((......))))))))))))(((((........))))))))... ( -15.50) >DroPse_CAF1 1303 112 + 1 CUCACCUCACGAAUCCACUCCAAUCCAAUCCAACUCAAAAUCGGUACGAACAUAC--UAUAGUACAACGGUUUCGAGCAACUGUGUAUUAACUUCACCAACGAGAAGUUGCAAC .......(((((......)).............(((.((((((((((........--....))))..)))))).))).....)))...(((((((........))))))).... ( -16.10) >DroSec_CAF1 1179 101 + 1 CUCAUCCCAUCAAUCCGCACCAUUUCCCC---ACAAAAA----------AUAUGAUAUCUAGUACAACGGUUUCGAGCAACUGUGUAUUAACUUCACCAACGAGAAGUUGCAAC ................(((..........---.......----------..........(((((((.(((((......))))))))))))(((((........))))))))... ( -15.50) >DroEre_CAF1 1175 99 + 1 CUCAUCCCAUCAAUCCGCACCAUUUCCCCC-----AAAA----------AUAUGACAUCUAGUACAACGGUUUCGAGCAACUGUGUAUUAACUUCACCAACGAGAAGUUGCAAC ................(((...........-----....----------..........(((((((.(((((......))))))))))))(((((........))))))))... ( -15.50) >DroWil_CAF1 1254 96 + 1 -AAAUCCUAUUAUUCC-CUCCACACC------ACCAAAU----------AUACCAAAUAUAGUACAACGGUUUCGAGCAACUGUGUAUUAACUUCACCAACGAGAAGUUGCAAC -...............-.........------.......----------..........(((((((.(((((......))))))))))))(((((........)))))...... ( -12.60) >DroAna_CAF1 1217 100 + 1 CUCAUCGCACCAAUCCUUGCCAAUUCCUA----CAAAAA----------AAAUGAUAUCUAGUACAACGGUUUCGAGCAACUGUGUAUUAACUUCACCAACGAGAAGUUGCAAC ......(((....................----......----------..........(((((((.(((((......))))))))))))(((((........))))))))... ( -15.50) >consensus CUCAUCCCAUCAAUCCGCACCAUUUCCCC___ACAAAAA__________AUAUGAUAUCUAGUACAACGGUUUCGAGCAACUGUGUAUUAACUUCACCAACGAGAAGUUGCAAC ............................................................((((((.(((((......)))))))))))((((((........))))))..... (-12.50 = -12.50 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:11 2006