| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,766,360 – 16,766,452 |
| Length | 92 |
| Max. P | 0.677977 |

| Location | 16,766,360 – 16,766,452 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -10.24 |
| Energy contribution | -10.11 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
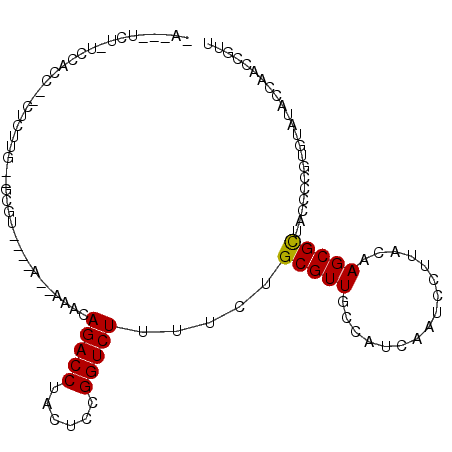
>2L_DroMel_CAF1 16766360 92 + 22407834 ---------CCAUCU--UUCUUG--GCCA----AAAAAACAGACCUACUCCGGUCUUUUCUGCGUUGCCAUCAAUCCUUACAAGCGUUACCCCGUGUAUACCAACCGUU ---------......--....((--((..----.......(((((......)))))..........)))).............(((......))).............. ( -11.71) >DroPse_CAF1 34692 100 + 1 CAUC-UCUAUCCACC--CUCUUGAUGCGU----G--AAACAGACCUACUCCGGUCUUUUCUGCGUUGCCAUCAAUCCUUACAAGCGCUACCCCGUGUAUACCAACCGUU ....-..........--...(((((((((----(--(((.(((((......))))))))).)))....)))))).........((((......))))............ ( -17.90) >DroGri_CAF1 34330 98 + 1 -A--UUCUUUCCACG--AUGAUG--GCAA----AAAAAAUAGACCUACUCUGGUCUUUUCUGCGUUGCCAUCAAUCCUUAUAAGCGCUACCCCGUCUAUACCAACCGUU -.--........(((--.(((((--((((----.......(((((......)))))........))))))))).....((((..((......))..)))).....))). ( -25.66) >DroWil_CAF1 47199 95 + 1 ---------UCAU-A--CUUUUCAUACAUUGACA--ACGUAGACCUACUCAGGUCUUUUCUGCGUUGCCAUCAAUCCUUACAAGCGCUACCCUGUGUAUACCAACCGUU ---------....-.--..........(((((((--((((((((((....)))))).....))))))...)))))........((((......))))............ ( -18.10) >DroMoj_CAF1 36451 99 + 1 -AUUUUCUCUCCGCUUCGUUAUG--GCGU-------AUAUAGACCUACUCCGGUCUUUUCUGCGUUGCCAUCAAUCCUUACAAGCGCUACCCCGUCUAUACUAACCGUU -..........(((((.((.(((--((((-------(...(((((......)))))....)))...)))))........)))))))....................... ( -16.70) >DroPer_CAF1 33070 100 + 1 CAUC-UCUAUCCACC--CUCUUGAUGCGU----G--AAACAGACCUACUCCGGUCUUUUCUGCGUUGCCAUCAAUCCUUACAAGCGCUACCCCGUGUAUACCAACCGUU ....-..........--...(((((((((----(--(((.(((((......))))))))).)))....)))))).........((((......))))............ ( -17.90) >consensus _A___UCU_UCCACC__CUCUUG__GCGU____A__AAACAGACCUACUCCGGUCUUUUCUGCGUUGCCAUCAAUCCUUACAAGCGCUACCCCGUGUAUACCAACCGUU ........................................(((((......))))).....(((((................)))))...................... (-10.24 = -10.11 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:07 2006