
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,765,428 – 16,765,565 |
| Length | 137 |
| Max. P | 0.954850 |

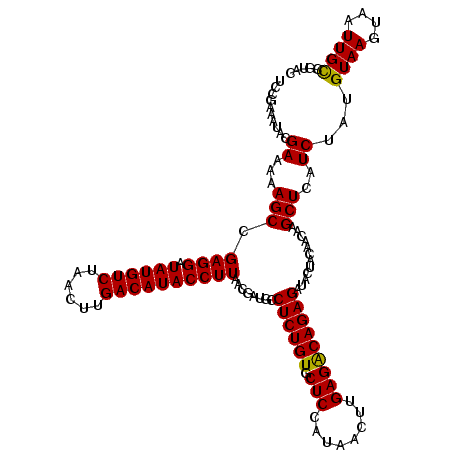
| Location | 16,765,428 – 16,765,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -24.70 |
| Energy contribution | -24.32 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
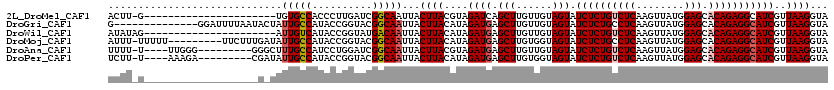
>2L_DroMel_CAF1 16765428 120 + 22407834 CCCGAAAUACGAAAAAGCCGAGGAUAUGUCCAACUUGACAUACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACAACAAGCUGAUCUACGUAAGUAAUUGCCGAUC ...............(((.((((.((((((......))))))))))........((((((.(((........)))))))))............)))((((...((((....)))).)))) ( -27.70) >DroVir_CAF1 35754 120 + 1 UCCGAAAUACGAAAAAGCUGAGGAUAUGUCUAACUUGACAUACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGGCAGAGAUAUUACAACAAGCUCAUCUAUGUAAGUAAUUGCCUUAC ..........((...((((((((.((((((......))))))))))..(..((((((.((........))..))))))..)...........))))..))...((((....))))..... ( -29.00) >DroGri_CAF1 33344 120 + 1 ACCGAAAUACGAAAAAGCUGAGGAUAUGUCUAACUUGACAUACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGGCAGAGAUACUACAACAAGCUCAUCUAUGUAAGUAAUUGCCGUAC .......((((....((((((((.((((((......))))))))))..(..((((((.((........))..))))))..)...........)))).......((((....)))))))). ( -31.70) >DroWil_CAF1 46153 120 + 1 UCCAAAAUACGAAAAAGCCGAGGAUAUGUCUAACUUGACAUACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACAACAAGCUCAUCUAUGUAAGUAAUUGUCAUAC ..........((...(((.((((.((((((......))))))))))........((((((.(((........)))))))))............)))..))(((((((....))).)))). ( -26.20) >DroMoj_CAF1 35529 120 + 1 ACCGAAAUACGAAAAAGCUGAGGAUAUGUCUAACUUGACAUACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGGCAGAGAUACUACCACAAGCUCAUCUAUGUAAGUAAUUGCCGUAC .......((((....((((((((.((((((......))))))))))..(..((((((.((........))..))))))..)...........)))).......((((....)))))))). ( -31.70) >DroPer_CAF1 32197 120 + 1 UCCAAAAUACGAAAAAGCCGAGGAUAUGUCCAACUUGACAUACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACCACAAGCUCAUCUAUGUAAGUAAUUGCCGUAC .......((((....(((.((((.((((((......))))))))))........((((((.(((........)))))))))............))).......((((....)))))))). ( -27.70) >consensus UCCGAAAUACGAAAAAGCCGAGGAUAUGUCUAACUUGACAUACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACAACAAGCUCAUCUAUGUAAGUAAUUGCCGUAC .......((((....(((.((((.((((((......))))))))))........((((((.(((........)))))))))............)))......)))).............. (-24.70 = -24.32 + -0.39)

| Location | 16,765,468 – 16,765,565 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.30 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16765468 97 + 22407834 UACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACAACAAGCUGAUCUACGUAAGUAAUUGCCGAUCAAGGGUGGCACA----------------------C-AAGU ((((((........((((((.(((........)))))))))..............(((((...((((....)))).))))))))))).....----------------------.-.... ( -23.10) >DroGri_CAF1 33384 106 + 1 UACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGGCAGAGAUACUACAACAAGCUCAUCUAUGUAAGUAAUUGCCGUACCGGUAUGGCAAUAGUAUUAAAAUCC--------------C ...........((((((.((........))..))))))..(((((((..((..((........))..))..((((((((....))))))))))))))).......--------------. ( -25.90) >DroWil_CAF1 46193 98 + 1 UACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACAACAAGCUCAUCUAUGUAAGUAAUUGUCAUACCGGUAUGACAAU----------------------CUAUAU ...((((..((((.((((((.(((........)))))))))...((......))..))))....))))..(((((((((....)))))))))----------------------...... ( -21.80) >DroMoj_CAF1 35569 110 + 1 UACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGGCAGAGAUACUACCACAAGCUCAUCUAUGUAAGUAAUUGCCGUACCGGUAUGGCAAUAUCAAAGAA---------AAAAA-AAAU ...((((....((((((.((........))..))))))(((.............))).......))))..(((((((((....))))))))).........---------.....-.... ( -24.02) >DroAna_CAF1 31010 106 + 1 UACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACAACAAGCUCAUCUACGUAAGUAAUUGCCGAUCCAGGAUGGCAAAGCCC---------CCCAA----A-AAAA ..............((((((.(((........)))))))))............(((....(((....))).(((((.(((...)))))))))))..---------.....----.-.... ( -21.30) >DroPer_CAF1 32237 106 + 1 UACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACCACAAGCUCAUCUAUGUAAGUAAUUGCCGUACCGGUAUGGCAAUAUCG---------UCUUU----A-AAGA .......(((((..((((((.(((........))))))))).((((...(((((.....)).))).))))(((((((((....)))))))))))))---------)....----.-.... ( -25.80) >consensus UACCUUAACGAUGCCUCUGUGCUCCAUAACUUGAGACAGAGAUACUACAACAAGCUCAUCUAUGUAAGUAAUUGCCGUACCGGUAUGGCAAUA_C___________________A_AAAU ...((((..((((.((((((.(((........)))))))))...((......))..))))....))))..(((((((((....)))))))))............................ (-21.38 = -21.30 + -0.08)

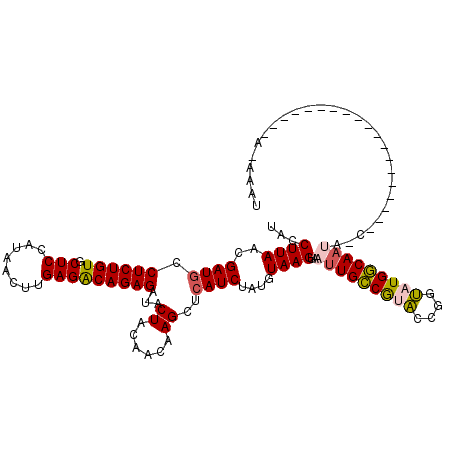

| Location | 16,765,468 – 16,765,565 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -19.81 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16765468 97 - 22407834 ACUU-G----------------------UGUGCCACCCUUGAUCGGCAAUUACUUACGUAGAUCAGCUUGUUGUAGUAUCUCUGUCUCAAGUUAUGGAGCACAGAGGCAUCGUUAAGGUA ....-.----------------------........(((((((..((.(((((..(((.((.....))))).)))))..(((((((((........))).))))))))...))))))).. ( -23.60) >DroGri_CAF1 33384 106 - 1 G--------------GGAUUUUAAUACUAUUGCCAUACCGGUACGGCAAUUACUUACAUAGAUGAGCUUGUUGUAGUAUCUCUGCCUCAAGUUAUGGAGCACAGAGGCAUCGUUAAGGUA .--------------..((((((((.....((((.....(((((.(((((..((((......))))...))))).)))))((((.(((........)))..))))))))..)))))))). ( -24.90) >DroWil_CAF1 46193 98 - 1 AUAUAG----------------------AUUGUCAUACCGGUAUGACAAUUACUUACAUAGAUGAGCUUGUUGUAGUAUCUCUGUCUCAAGUUAUGGAGCACAGAGGCAUCGUUAAGGUA .....(----------------------(((((((((....)))))))))).((((....((((.(((......))).((((((((((........))).)))))))))))..))))... ( -28.80) >DroMoj_CAF1 35569 110 - 1 AUUU-UUUUU---------UUCUUUGAUAUUGCCAUACCGGUACGGCAAUUACUUACAUAGAUGAGCUUGUGGUAGUAUCUCUGCCUCAAGUUAUGGAGCACAGAGGCAUCGUUAAGGUA ....-.....---------..(((((((((((((.((....)).))))))..........((((((((((.(((((.....))))).)))))).((.....))....))))))))))).. ( -28.20) >DroAna_CAF1 31010 106 - 1 UUUU-U----UUGGG---------GGGCUUUGCCAUCCUGGAUCGGCAAUUACUUACGUAGAUGAGCUUGUUGUAGUAUCUCUGUCUCAAGUUAUGGAGCACAGAGGCAUCGUUAAGGUA ....-(----(..((---------((((...))).)))..))..........((((....((((.(((......))).((((((((((........))).)))))))))))..))))... ( -27.10) >DroPer_CAF1 32237 106 - 1 UCUU-U----AAAGA---------CGAUAUUGCCAUACCGGUACGGCAAUUACUUACAUAGAUGAGCUUGUGGUAGUAUCUCUGUCUCAAGUUAUGGAGCACAGAGGCAUCGUUAAGGUA ....-.----...((---------((((((((((.((....)).))))))((((..(((((......)))))..))))((((((((((........))).))))))).))))))...... ( -28.70) >consensus AUUU_U___________________G_UAUUGCCAUACCGGUACGGCAAUUACUUACAUAGAUGAGCUUGUUGUAGUAUCUCUGUCUCAAGUUAUGGAGCACAGAGGCAUCGUUAAGGUA .............................(((((..........)))))...((((....((((.(((......))).((((((((((........))).)))))))))))..))))... (-19.81 = -20.20 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:06 2006