
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,754,238 – 16,754,338 |
| Length | 100 |
| Max. P | 0.514724 |

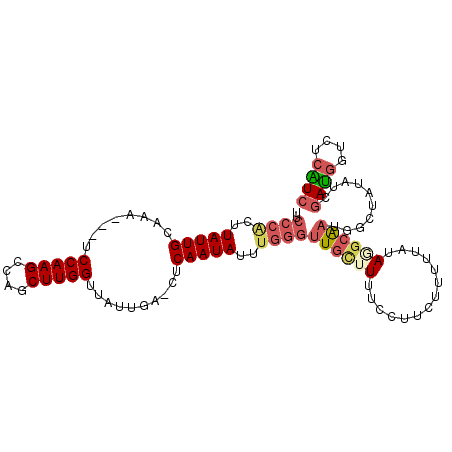
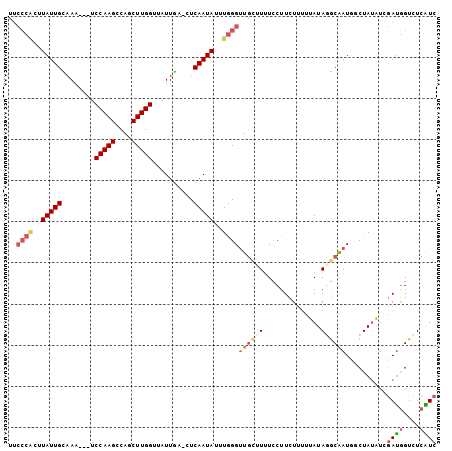
| Location | 16,754,238 – 16,754,338 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -13.22 |
| Energy contribution | -14.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
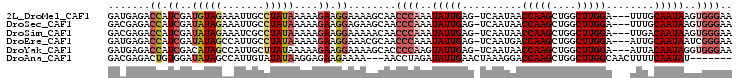
>2L_DroMel_CAF1 16754238 100 + 22407834 UUCCCACUUAUUGCAAA---UCCAAGCCAGCUUGGUUAUUGA-CUCAAUAUUUGGGUUGCUUUUCCUUCUUUUUAUAGGCAAUUUCUACAUCGAUGGUCUCAUC ..((((..((((((((.---.(((((....)))))...))).-..)))))..))))((((((..............))))))..........((((....)))) ( -20.94) >DroSec_CAF1 17629 100 + 1 UUCCCACUUAUUGCAAA---UCCAAGCCAGCUUGGUUAUUGA-CUCAAUAUUUGGGUUGCUUCUCCUUCUUUUUAUAGGCAAUUUCUAUAUCGAUGGUCUCGUC ..((((..((((((((.---.(((((....)))))...))).-..)))))..))))........((.((....((((((.....))))))..)).))....... ( -20.30) >DroSim_CAF1 17627 100 + 1 UUCCCACUUAUUGUCAA---UCCAAGCCAGCUUGGUUAUUGA-CUCAAUAUUUGGGUUGUUUUUCCUUCUUUUUAUAGGCGAUUUCUAUAUCGAUGGUCUCGUC ..((((..(((((((((---((((((....)))))..)))))-)..))))..)))).....................(((((...((((....))))..))))) ( -24.50) >DroEre_CAF1 17675 100 + 1 UUCCCGAUUAUUGCAAU---UCCAAGCCAGCUUGGUCAUUGA-CUCAAUAUUUGGGUUGCGUUUCCUUCUUUUUAUAGGCAAUGGCUAUAUCGAUGGUCUCAUC ..(((((.(((((((((---.(((((....)))))..)))).-..))))).)))))((((.(..............).)))).((((((....))))))..... ( -23.84) >DroYak_CAF1 17624 100 + 1 UUCCCACCUAUUGUAAU---UCCAAGCCAGCUUGGUUAUUGA-CUCAAUACUUGGGGUGCUUUUCCUUCUUUUUAUAAGCAAUGGCUAUGUCGAUGGUCUCAUC .(((((..(((((((((---.(((((....)))))..)))).-..)))))..)))))(((((..............)))))..((((((....))))))..... ( -23.64) >DroAna_CAF1 19381 94 + 1 -------AUAUUGAAAAGUUGCCAAGCCAGCUUGGUCCUUUAGUUCAAUAUCUAGGUU---UUUUCUUCUCCUUAUAUACAAUGGCUAUAUCCACAGUCUCGUC -------((((((((.....((((((....)))))).......))))))))..(((..---.........)))..........((((........))))..... ( -15.20) >consensus UUCCCACUUAUUGCAAA___UCCAAGCCAGCUUGGUUAUUGA_CUCAAUAUUUGGGUUGCUUUUCCUUCUUUUUAUAGGCAAUGGCUAUAUCGAUGGUCUCAUC ..((((..(((((........(((((....)))))..........)))))..))))((((((..............))))))..........((((....)))) (-13.22 = -14.23 + 1.00)

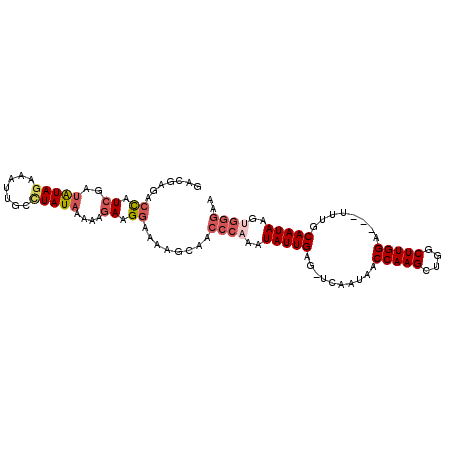
| Location | 16,754,238 – 16,754,338 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -14.01 |
| Energy contribution | -15.12 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16754238 100 - 22407834 GAUGAGACCAUCGAUGUAGAAAUUGCCUAUAAAAAGAAGGAAAAGCAACCCAAAUAUUGAG-UCAAUAACCAAGCUGGCUUGGA---UUUGCAAUAAGUGGGAA ((((....))))..(((((.......))))).................((((..(((((..-.(((...(((((....))))).---.))))))))..)))).. ( -22.10) >DroSec_CAF1 17629 100 - 1 GACGAGACCAUCGAUAUAGAAAUUGCCUAUAAAAAGAAGGAGAAGCAACCCAAAUAUUGAG-UCAAUAACCAAGCUGGCUUGGA---UUUGCAAUAAGUGGGAA .......((.((..(((((.......)))))....)).))........((((..(((((..-.(((...(((((....))))).---.))))))))..)))).. ( -21.90) >DroSim_CAF1 17627 100 - 1 GACGAGACCAUCGAUAUAGAAAUCGCCUAUAAAAAGAAGGAAAAACAACCCAAAUAUUGAG-UCAAUAACCAAGCUGGCUUGGA---UUGACAAUAAGUGGGAA .......((.((..(((((.......)))))....)).))........((((..((((..(-(((((..(((((....))))))---)))))))))..)))).. ( -24.40) >DroEre_CAF1 17675 100 - 1 GAUGAGACCAUCGAUAUAGCCAUUGCCUAUAAAAAGAAGGAAACGCAACCCAAAUAUUGAG-UCAAUGACCAAGCUGGCUUGGA---AUUGCAAUAAUCGGGAA ((((....))))..........((((((......))..(....)))))(((...(((((..-.((((..(((((....))))).---)))))))))...))).. ( -21.90) >DroYak_CAF1 17624 100 - 1 GAUGAGACCAUCGACAUAGCCAUUGCUUAUAAAAAGAAGGAAAAGCACCCCAAGUAUUGAG-UCAAUAACCAAGCUGGCUUGGA---AUUACAAUAGGUGGGAA ((((....))))......((..((.(((........))).))..))..((((..(((((((-(......(((((....))))).---))).)))))..)))).. ( -21.60) >DroAna_CAF1 19381 94 - 1 GACGAGACUGUGGAUAUAGCCAUUGUAUAUAAGGAGAAGAAAA---AACCUAGAUAUUGAACUAAAGGACCAAGCUGGCUUGGCAACUUUUCAAUAU------- ......((.((((......)))).)).......((((((....---..(((((........))..))).(((((....)))))...)))))).....------- ( -17.30) >consensus GACGAGACCAUCGAUAUAGAAAUUGCCUAUAAAAAGAAGGAAAAGCAACCCAAAUAUUGAG_UCAAUAACCAAGCUGGCUUGGA___UUUGCAAUAAGUGGGAA .......((.((..(((((.......)))))....)).))........((((..(((((..........(((((....)))))........)))))..)))).. (-14.01 = -15.12 + 1.11)
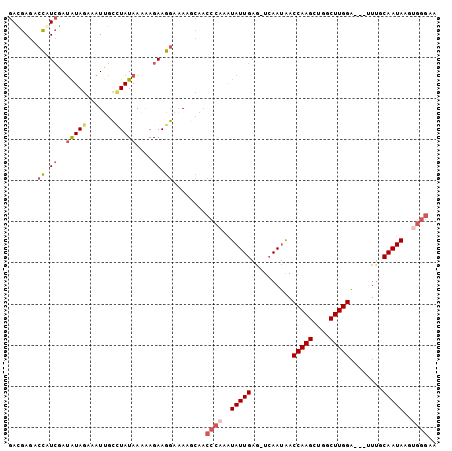
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:58 2006