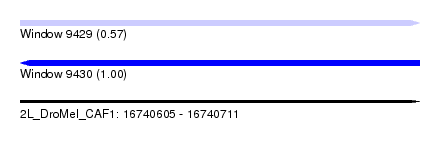
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,740,605 – 16,740,711 |
| Length | 106 |
| Max. P | 0.996415 |

| Location | 16,740,605 – 16,740,711 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16740605 106 + 22407834 CUUUU-UGCUAAAGUAAUGCUACCAUAAGUGCUCCGCCUACCACCAAUCAUGCUGGUGGCGCAUCCAGCGCACCACCAAGUGGUCCGCCAGAUGCUCAGCAACACCA ....(-(((....))))((((..............(((.((((.((....)).)))))))(((((..(((.(((((...))))).)))..)))))..))))...... ( -33.40) >DroPse_CAF1 6241 106 + 1 UUUUU-UGGUAAAGUAAUGCUACCAUAAGUGCUCCACCCACCACCAAUCAUGCUGGUGGCGCAUCAAGCGCACCACCAAGUGGCCCGCCCGAUGCACAACAGCAACA .....-(((((.(....)..)))))....((((....((((((.((....)).)))))).(((((..(((..((((...))))..)))..))))).....))))... ( -33.10) >DroEre_CAF1 4234 106 + 1 CUUUU-UGCUAAAGUAAUGCUACCAUAAGUGCUCCGCCAACCACCAAUCAUGCUGGUGGCACAUCCAGCGCGCCACCAAGUGGUCCGCCCGAUGCUCAGCAACACCA ....(-((((.......((((((((...(((.................)))..))))))))((((..(((.(((((...))))).)))..))))...)))))..... ( -29.63) >DroYak_CAF1 4253 106 + 1 CUUUU-UACUAAAGUAAUGCUACCAUAAGUGCUCCGCCUACCACCAAUCAUGCUGGUGGCGCAUCCAGUGCACCACCAAGUGGCCCGCCCGAUGCUCAGCAACACCA ....(-(((....))))((((..............(((.((((.((....)).)))))))(((((..(((..((((...))))..)))..)))))..))))...... ( -29.10) >DroAna_CAF1 3708 107 + 1 AUUCCCCCAUGUAGUAAUGCAACCAUAAGUGCUCCGCCCACCACCAACCAUGCUGGUGGCGUACCUGGCGCACCACCAAGUGGCCCGCCAGAUGCCCAGCAACCACC .........((((....))))........((((..(((.((((.((....)).)))))))(((.((((((..((((...))))..)))))).)))..))))...... ( -34.60) >DroPer_CAF1 4318 106 + 1 UUUUU-UGGUAAAGUAAUGCUACCAUAAGUGCUCCACCCACCACCAAUCAUGCUGGUGGCGCAUCAAGCGCACCACCAAGUGGCCCGCCCGAUGCACAACAGCAACA .....-(((((.(....)..)))))....((((....((((((.((....)).)))))).(((((..(((..((((...))))..)))..))))).....))))... ( -33.10) >consensus CUUUU_UGCUAAAGUAAUGCUACCAUAAGUGCUCCGCCCACCACCAAUCAUGCUGGUGGCGCAUCCAGCGCACCACCAAGUGGCCCGCCCGAUGCUCAGCAACAACA .............((...(((((((...(((.................)))..)))))))(((((..(((..((((...))))..)))..)))))...))....... (-23.68 = -23.68 + 0.00)

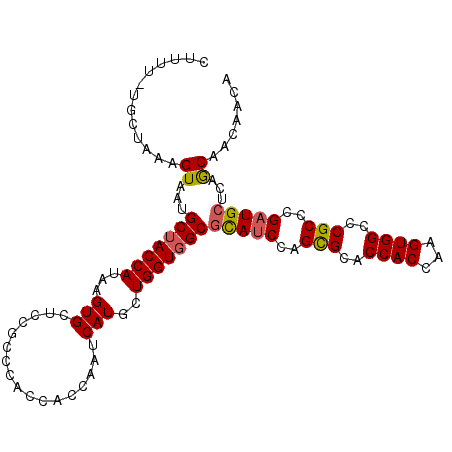
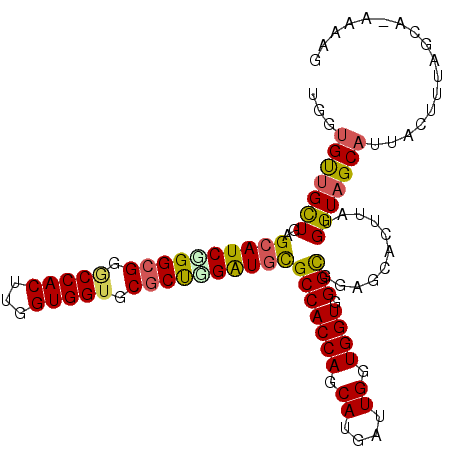
| Location | 16,740,605 – 16,740,711 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -46.37 |
| Consensus MFE | -40.52 |
| Energy contribution | -40.25 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16740605 106 - 22407834 UGGUGUUGCUGAGCAUCUGGCGGACCACUUGGUGGUGCGCUGGAUGCGCCACCAGCAUGAUUGGUGGUAGGCGGAGCACUUAUGGUAGCAUUACUUUAGCA-AAAAG ((((((((((..(((((..(((.(((((...))))).)))..)))))(((((((.((....)).)))).)))...........))))))))))........-..... ( -48.70) >DroPse_CAF1 6241 106 - 1 UGUUGCUGUUGUGCAUCGGGCGGGCCACUUGGUGGUGCGCUUGAUGCGCCACCAGCAUGAUUGGUGGUGGGUGGAGCACUUAUGGUAGCAUUACUUUACCA-AAAAA (((((((((.((((((((((((.(((((...))))).))))))))((.((((((.((....)).))))))))...))))..)))))))))...........-..... ( -48.20) >DroEre_CAF1 4234 106 - 1 UGGUGUUGCUGAGCAUCGGGCGGACCACUUGGUGGCGCGCUGGAUGUGCCACCAGCAUGAUUGGUGGUUGGCGGAGCACUUAUGGUAGCAUUACUUUAGCA-AAAAG ((((((((((..(((((.(((((.((((...))))).)))).)))))(((((((.((....)).))).))))...........))))))))))........-..... ( -40.90) >DroYak_CAF1 4253 106 - 1 UGGUGUUGCUGAGCAUCGGGCGGGCCACUUGGUGGUGCACUGGAUGCGCCACCAGCAUGAUUGGUGGUAGGCGGAGCACUUAUGGUAGCAUUACUUUAGUA-AAAAG ((((((((((..(((((.((.(.(((((...))))).).)).)))))(((((((.((....)).)))).)))...........))))))))))........-..... ( -42.50) >DroAna_CAF1 3708 107 - 1 GGUGGUUGCUGGGCAUCUGGCGGGCCACUUGGUGGUGCGCCAGGUACGCCACCAGCAUGGUUGGUGGUGGGCGGAGCACUUAUGGUUGCAUUACUACAUGGGGGAAU ......((((..((((((((((.(((((...))))).)))))))).((((((((((...)))))))))).))..))))((((((............))))))..... ( -49.70) >DroPer_CAF1 4318 106 - 1 UGUUGCUGUUGUGCAUCGGGCGGGCCACUUGGUGGUGCGCUUGAUGCGCCACCAGCAUGAUUGGUGGUGGGUGGAGCACUUAUGGUAGCAUUACUUUACCA-AAAAA (((((((((.((((((((((((.(((((...))))).))))))))((.((((((.((....)).))))))))...))))..)))))))))...........-..... ( -48.20) >consensus UGGUGUUGCUGAGCAUCGGGCGGGCCACUUGGUGGUGCGCUGGAUGCGCCACCAGCAUGAUUGGUGGUGGGCGGAGCACUUAUGGUAGCAUUACUUUAGCA_AAAAG ...(((((((..((((((((((.(((((...))))).))))))))))(((((((.((....)).)))).)))...........)))))))................. (-40.52 = -40.25 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:53 2006