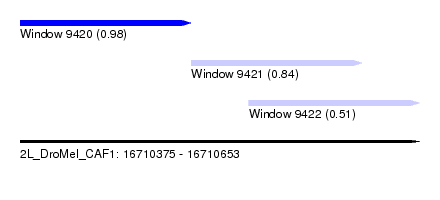
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,710,375 – 16,710,653 |
| Length | 278 |
| Max. P | 0.977209 |

| Location | 16,710,375 – 16,710,494 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -28.74 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16710375 119 + 22407834 UUAUGUCUCUGCCAGCAGGACAAAAGCAAAUUAAUUAAAUUAUAGCAAUUCCUC-AACACUUAUGCUGUAAUUGAUGUUGGGGCACCAACACCAUGUGAACUAGCUGCAGCUCAUAACAA (((((...((((.(((((..((...............((((((((((.......-........))))))))))(.((((((....)))))).)...))..)).)))))))..)))))... ( -30.16) >DroSec_CAF1 164970 119 + 1 UUAUGUCUCUGCCAGCAGGACAA-AGCAAAUUAAUUAAAUUAUAGCAAUUCCUGUUACACUUAUGCUGUAAUUGCUGUUGGGGCACCAACACCAUGUGAACUAGCUGCAGCUCAUAACGA (((((...((((.(((((..((.-.............((((((((((................))))))))))(.((((((....)))))).)...))..)).)))))))..)))))... ( -30.09) >DroEre_CAF1 166244 117 + 1 UUAUGUCUCUGCCAGCAGGACAA-AGCAAAUUAAUUAAAUUAUAGCAAUUCCUC-AACAUUUAUGCUGUAAUUGAUGUUGGGCCACC-ACAGCAUGUGAACUAGCUGCGGCUCAUAACAA ...((((.(((....))))))).-.............((((((((((.......-........))))))))))..(((((((((...-.((((..(....)..)))).))))))..))). ( -30.06) >consensus UUAUGUCUCUGCCAGCAGGACAA_AGCAAAUUAAUUAAAUUAUAGCAAUUCCUC_AACACUUAUGCUGUAAUUGAUGUUGGGGCACCAACACCAUGUGAACUAGCUGCAGCUCAUAACAA (((((...((((.(((((..((...............((((((((((................))))))))))(.((((((....)))))).)...))..)).)))))))..)))))... (-28.74 = -28.86 + 0.11)

| Location | 16,710,494 – 16,710,613 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -29.78 |
| Energy contribution | -30.03 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16710494 119 + 22407834 AAGGCAUACGGCUCAUUGGCCUAUAUAAAAUGCCAGUGCCAUGCCAAGGCAUAAACAA-AAGGCAGAAAUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUG ..(((((..((((....))))........))))).(((((((((....))))......-....((....))..............(((........))))))))((((((...)))))). ( -33.70) >DroSec_CAF1 165089 119 + 1 AAGGCAUACGGCUCAUUGGCCUAUAUAAAAUGCCAGUGCCAUGCCAAGGCAUAAACAA-AAGGCAGAAAUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUG ..(((((..((((....))))........))))).(((((((((....))))......-....((....))..............(((........))))))))((((((...)))))). ( -33.70) >DroEre_CAF1 166361 110 + 1 AAGGCAUACGGCUCAUUGGCCUAUAUAAAAUGCCAGUGCCA----------UAAACAAAAAGGCAGAAGUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUG ..(((((..((((....))))........))))).(((((.----------................(((.........)))...(((........))))))))((((((...)))))). ( -28.60) >consensus AAGGCAUACGGCUCAUUGGCCUAUAUAAAAUGCCAGUGCCAUGCCAAGGCAUAAACAA_AAGGCAGAAAUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUG ..(((((..((((....))))........))))).(((((((((....)))).................................(((........))))))))((((((...)))))). (-29.78 = -30.03 + 0.25)

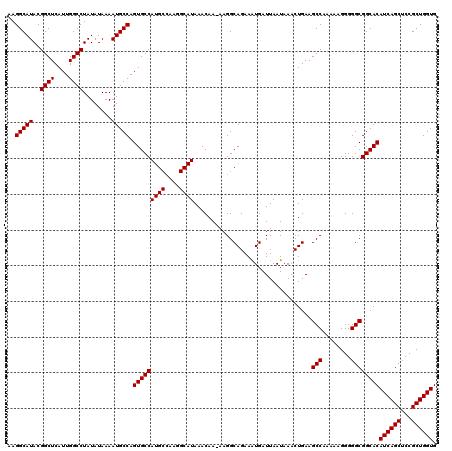
| Location | 16,710,534 – 16,710,653 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.76 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.33 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16710534 119 + 22407834 AUGCCAAGGCAUAAACAA-AAGGCAGAAAUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUGUGUGGAAUACAGAUGAUGUGACAUAAUUGAAAAACCGGAG ((((....))))......-..((((....)).(((((...(((..(((........))).((((((((((...))))))))))......)))(((......))).)))))....)).... ( -28.30) >DroSec_CAF1 165129 119 + 1 AUGCCAAGGCAUAAACAA-AAGGCAGAAAUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUGUGUGGAAUACAGAUGAUGUGACAUAAUUGAAAAACCGGAG ((((....))))......-..((((....)).(((((...(((..(((........))).((((((((((...))))))))))......)))(((......))).)))))....)).... ( -28.30) >DroEre_CAF1 166401 110 + 1 A----------UAAACAAAAAGGCAGAAGUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUGUGUGGAAUACAGAUGAUGUGACAUAAUUGAAAAUCCGGAG .----------..........(((...(((.........)))...))).....(((..(.((((((((((...)))))))))).)..((((.....)))).............))).... ( -26.40) >consensus AUGCCAAGGCAUAAACAA_AAGGCAGAAAUGAUUAAUAAACUGAAGCCAAAAAGGGGGCGGCACAUCAGCUCCGCUGGUGUGUGGAAUACAGAUGAUGUGACAUAAUUGAAAAACCGGAG ((((....))))...........(((..(((.(((.....(((..(((........))).((((((((((...))))))))))......)))......))))))..)))........... (-23.30 = -23.33 + 0.03)

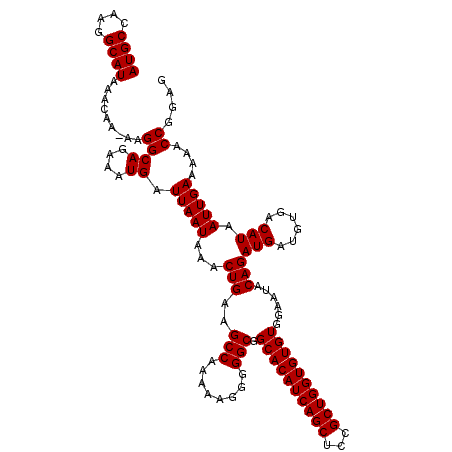
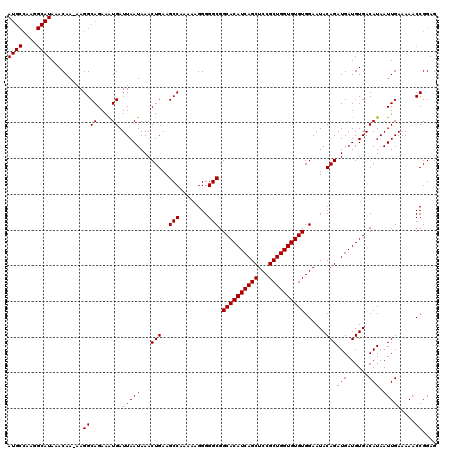
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:45 2006