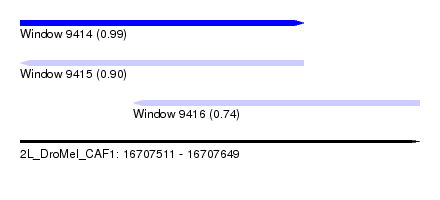
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,707,511 – 16,707,649 |
| Length | 138 |
| Max. P | 0.990811 |

| Location | 16,707,511 – 16,707,609 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -25.73 |
| Energy contribution | -24.63 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
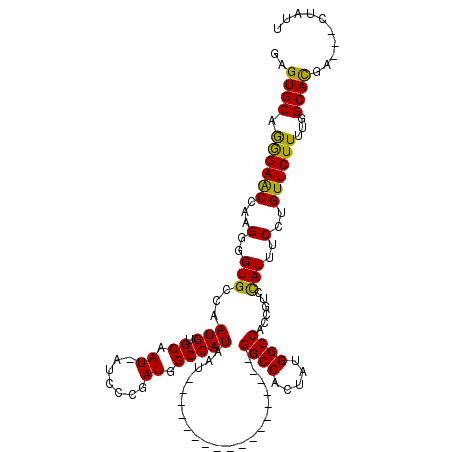
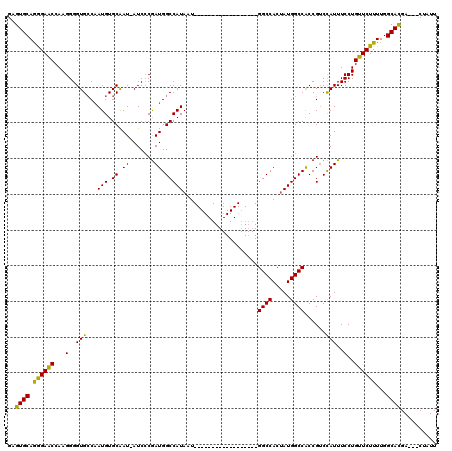
>2L_DroMel_CAF1 16707511 98 + 22407834 GAGUGCAGGGAACCAAGGGGUGCCAAUGUGCAAU-AUCCCGAUGGCCAUAAU------------------GGCCACUAUGGCCACCGUCCAUUUCCUGUUCUUUUGGCACGA---CUAUU ..((((.((((((...(((((((......)))..-.))))(((((......(------------------((((.....))))))))))........))))))...))))..---..... ( -31.30) >DroSec_CAF1 162100 99 + 1 GAGUGCAGGGAGCCAAGGGGUGCCAAUGUGCGAUAAUCCCGAUGGCCAUAAU------------------GGCCACUAUGGCCGCCGUCCAUUUCCUGUUCUUUUGGCACGA---CUAUU ..((((.((((((...(((((((......))....)))))((((((......------------------((((.....))))))))))........))))))...))))..---..... ( -32.50) >DroEre_CAF1 163252 119 + 1 GAGUGCAAAGAACCAAGGGGUGGCCAUGUGCAAU-AUCACAAUGGCCAUAAUGGCCACAAUGGUCAUAAUGGCCACAAUGGCCACCGACUAUUUCCUGUUCUUUUGGCAUGAUUACUAUU ..((((.((((((..(((((((((((((((....-..)))..(((((((.((((((.....)))))).)))))))..)))))))))........)))))))))...)))).......... ( -49.40) >consensus GAGUGCAGGGAACCAAGGGGUGCCAAUGUGCAAU_AUCCCGAUGGCCAUAAU__________________GGCCACUAUGGCCACCGUCCAUUUCCUGUUCUUUUGGCACGA___CUAUU ..((((.((((((...(..(((...(((.((.((.......)).))))).....................((((.....))))......)))..)..))))))...)))).......... (-25.73 = -24.63 + -1.10)

| Location | 16,707,511 – 16,707,609 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -35.91 |
| Consensus MFE | -21.94 |
| Energy contribution | -23.38 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16707511 98 - 22407834 AAUAG---UCGUGCCAAAAGAACAGGAAAUGGACGGUGGCCAUAGUGGCC------------------AUUAUGGCCAUCGGGAU-AUUGCACAUUGGCACCCCUUGGUUCCCUGCACUC .....---..((((.....(((((((.((((..(((((((((((((....------------------)))))))))))))...)-)))((......))...)))..))))...)))).. ( -33.60) >DroSec_CAF1 162100 99 - 1 AAUAG---UCGUGCCAAAAGAACAGGAAAUGGACGGCGGCCAUAGUGGCC------------------AUUAUGGCCAUCGGGAUUAUCGCACAUUGGCACCCCUUGGCUCCCUGCACUC ....(---((((.(((.............))))))))(((((((((....------------------)))))))))...((((.....((.((..((...))..))))))))....... ( -29.72) >DroEre_CAF1 163252 119 - 1 AAUAGUAAUCAUGCCAAAAGAACAGGAAAUAGUCGGUGGCCAUUGUGGCCAUUAUGACCAUUGUGGCCAUUAUGGCCAUUGUGAU-AUUGCACAUGGCCACCCCUUGGUUCUUUGCACUC ...........(((..((((((((((........((((((((..((((((((.(((.((.....)).))).))))))))((((..-....)))))))))))))))..))))))))))... ( -44.40) >consensus AAUAG___UCGUGCCAAAAGAACAGGAAAUGGACGGUGGCCAUAGUGGCC__________________AUUAUGGCCAUCGGGAU_AUUGCACAUUGGCACCCCUUGGUUCCCUGCACUC ..........((((.....(((((((.......(((((((((((((......................)))))))))))))........((......))...)))..))))...)))).. (-21.94 = -23.38 + 1.45)

| Location | 16,707,550 – 16,707,649 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.48 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
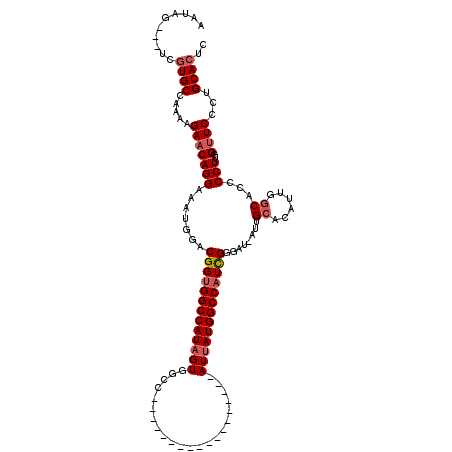
>2L_DroMel_CAF1 16707550 99 - 22407834 UAUAAUAUUUGCAUUUGUAUUGGCUACGUGGCGUAUGCGUAAUAG---UCGUGCCAAAAGAACAGGAAAUGGACGGUGGCCAUAGUGGCC------------------AUUAUGGCCAUC .....(((((.(..((.(.(((((.(((..(((....))).....---.)))))))).).))..).)))))...((((((((((((....------------------)))))))))))) ( -31.10) >DroSec_CAF1 162140 99 - 1 UAUAAUAUUUGCAUUUGUAUUGGCUACGUGGCGUAUGCGUAAUAG---UCGUGCCAAAAGAACAGGAAAUGGACGGCGGCCAUAGUGGCC------------------AUUAUGGCCAUC .....(((((((((.(((..((....))..))).))))).))))(---((((.(((.............))))))))(((((((((....------------------)))))))))... ( -29.52) >DroEre_CAF1 163291 120 - 1 UAUAAUAUUUGCAUUUGUAUUGGGUACGUGGCGUAUGCGUAAUAGUAAUCAUGCCAAAAGAACAGGAAAUAGUCGGUGGCCAUUGUGGCCAUUAUGACCAUUGUGGCCAUUAUGGCCAUU .....(((((.(....((((...)))).((((((.(((......)))...))))))........).)))))...(((((((((.((((((((.((....)).)))))))).))))))))) ( -39.10) >consensus UAUAAUAUUUGCAUUUGUAUUGGCUACGUGGCGUAUGCGUAAUAG___UCGUGCCAAAAGAACAGGAAAUGGACGGUGGCCAUAGUGGCC__________________AUUAUGGCCAUC .....(((((.(..((.(.(((((((((((.....))))))...........))))).).))..).)))))...((((((((((((......................)))))))))))) (-21.19 = -21.75 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:39 2006